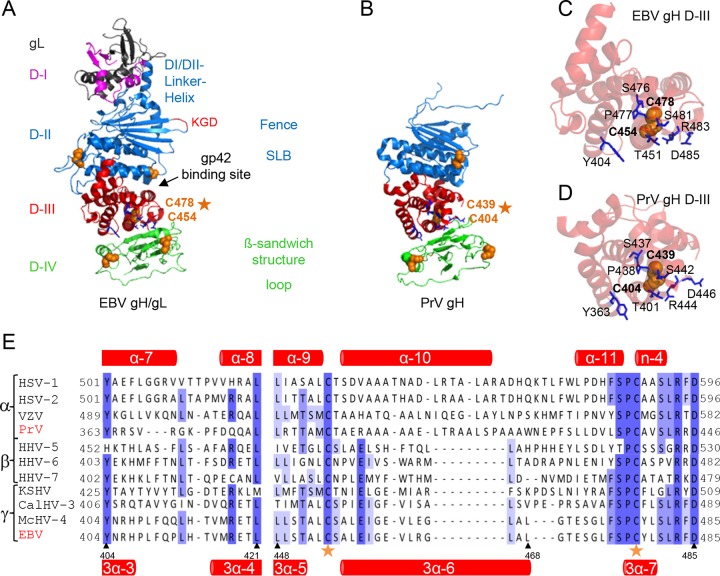
FIG 1.
Side-by-side comparison of EBV gH/gL and PrV gH. The EBV gH/gL (PDB code 3PHF) (A) and PrV gH (PDB code 2XQY) (B) structures (4, 6) are shown as ribbon diagrams. The four domains (D) are indicated in different colors. Previously identified structural aspects, such as the bifunctional KGD motif (red) (24), DBs (orange spheres), and syntaxin-like bundle (SLB) formed by three helices, are shown. The structural views of D-III of EBV gH/gL (C) and PrV gH (D), including the conserved DB (orange spheres) and conserved surrounding amino acids (blue sticks), are shown as ribbon diagrams. (E) Amino acid alignment of the conserved DB (orange stars) of a partial D-III (corresponding to amino acids 404 to 421 and 448 to 485 of EBV gH) is shown with the correlated secondary structures, such as alpha-helices and β-sheets (red) for PrV (top) and EBV (bottom) gH.