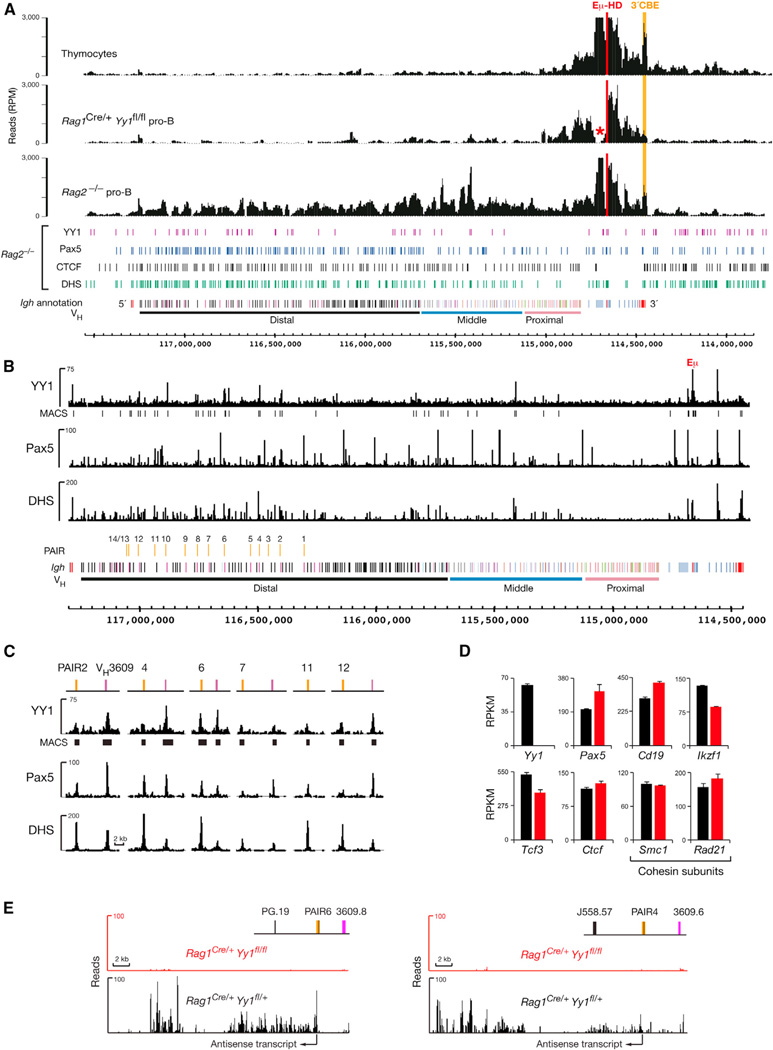
Figure 7. YY1 Controls PAIR Function and Long-Range Interactions across the Igh Locus.
(A) 4C-seq analysis of ex vivo sorted Rag1Cre/+Yy1fl/fl pro-B cells (Figures S6B and S6C), control Rag2−/− pro-B cells, and thymocytes with the viewpoint Eµ-HD (red). The Eµ-3′CBE interaction is highlighted in orange. A red asterisk indicates the loss of DH sequences by DH-JH rearrangements in the recombination-proficient Rag1Cre/+Yy1fl/fl pro-B cells.
(B) Identification of YY1 peaks in the Igh locus by Bio-ChIP-sequencing of cultured pro-B cells from Yy1ihCd2/+Rosa26BirA/BirARag2−/− mice. Vertical bars below the Bio-ChIP-seq track identify significant YY1 peaks that were called by MACS with a p value of <10−10. For comparison, the Pax5-binding and DHS site patterns of Rag2−/− pro-B cells are shown (Revilla-I-Domingo et al., 2012).
(C) YY1 binding at PAIR elements and associated VH3609 genes.
(D and E) Analysis of YY1-dependent gene expression. The expression values (RPKM) of selected genes (D) and RNA-seq profiles of the antisense transcripts originating from PAIR4 and PAIR6 (E) are shown for Rag1Cre/+Yy1fl/fl (red) and control Rag1Cre/+Yy1fl/+ (black) pro-B cells together with the standard error of the mean (based on two RNA-seq experiments for each genotype).
See also Figure S6.