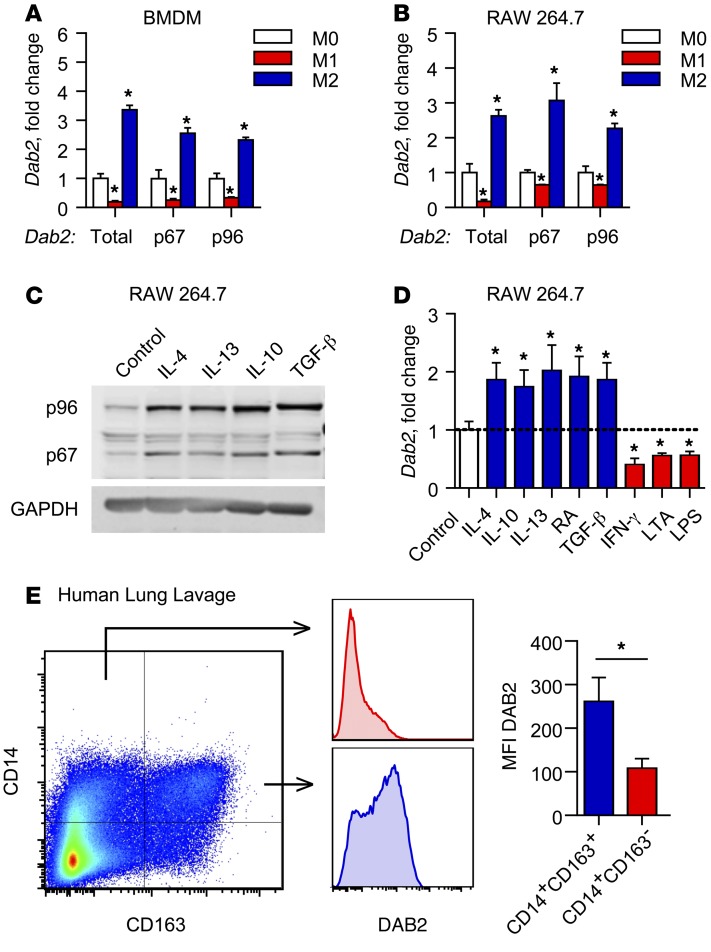
Figure 1. Dab2 expression is differentially regulated in M1 and M2 macrophages.
mRNA expression of total Dab2 and p67 and p96 Dab2 splice variants was determined by qPCR of BMDMs (A) and RAW 264.7 macrophages (B). For A and B, cells were treated with control media (M0), 1 μg/ml LPS and 10 U/ml IFN-γ (M1), or 10 ng/ml IL-4 (M2) for 3 hours. Results were normalized to B2m (encoding β-2-microglobulin) mRNA and are presented as fold change relative to M0. (A and B) Data are representative of 3 independent experiments. *P < 0.0001, by 2-way ANOVA with Dunnett’s multiple comparisons test. (C) Western blot analysis of p96-DAB2 and p67-DAB2 protein in RAW 264.7 macrophages treated for 24 hours with control media, 10 ng/ml IL-4, 10 ng/ml IL-13, 10 ng/ml IL-10, or 5 ng/ml TGF-β. GAPDH was used as a loading control. (D) qPCR analysis of total Dab2 mRNA expression in RAW 264.7 macrophages treated for 3 hours with control media (M0), 10 ng/ml IL-4, 10 ng/ml IL-10, 10 ng/ml IL-13, 10 μM 9-cis-retinoic acid (RA), 5 ng/ml TGF-β, 10 U/ml IFN-γ, 1 μg/ml LPS, or 1 μg/ml LTA. Results are normalized to B2m mRNA and are presented relative to M0. Data were compiled from 2 independent experiments and are presented as the mean ± SEM of quadruplicate treatments. *P < 0.05, by Student’s t test or Mann-Whitney U test. (E) DAB2 was preferentially expressed in M2-like human macrophages that expressed CD163. BAL fluid obtained from critically ill patients was analyzed by flow cytometry for CD14 and the macrophage M2 marker CD163. DAB2 MFI was determined for CD14+CD163– and CD14+CD163+ cells (bar graph shows the mean ± SEM, n = 6). *P < 0.02, by 2-tailed, paired Student’s t test.