Fig. 4. Self-renewal genes are associated with distinct enhancers in ES cells and macrophages.
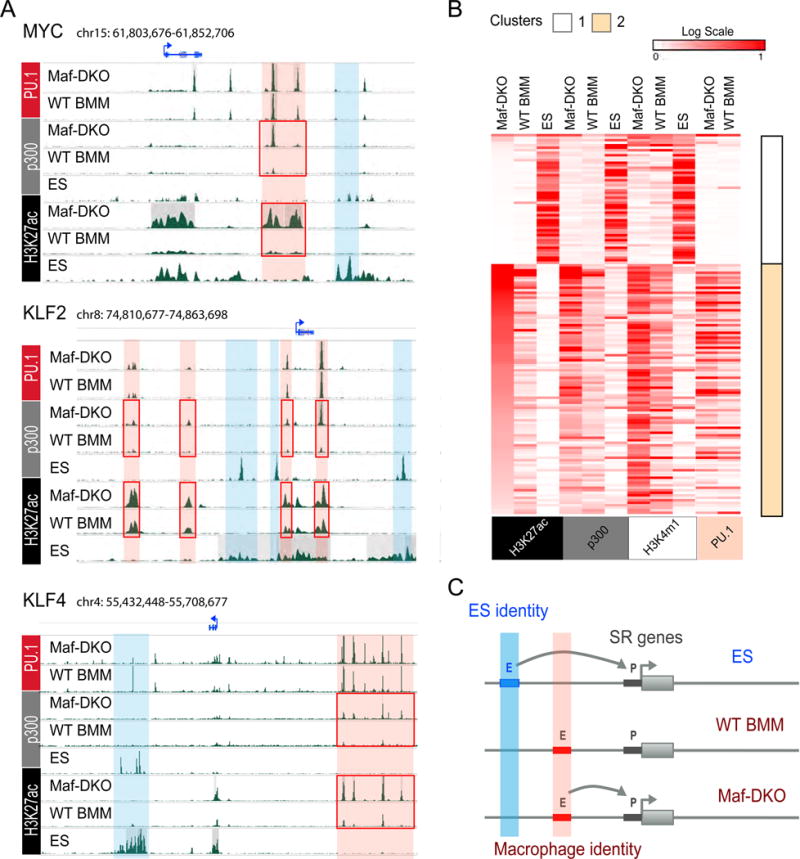
A) Genomic regions surrounding MYC, KLF2 and KLF4 genes showing distinct ES cell (blue) and macrophage (red) specific predicted enhancer regions with differential H3K27ac and p300 enrichment in Maf-DKO over WT BMM (red boxes). B) Heatmaps and k-means clustering (k=2) of Chip-seq signal of all H3K27ac+ regions associated with self-renewal genes in ES cells, Maf-DKO and WT BMM, including both differentially regulated Maf-DKO-only and non-differentially regulated regions. Corresponding regions are shown for p300, H3K4m1 and PU.1 (ES, Maf-DKO and WT BMM). C) Model based on panels A) and B) to describe tissue-specific macrophage and ES cell enhancer platforms associated with individual self-renewal genes.
