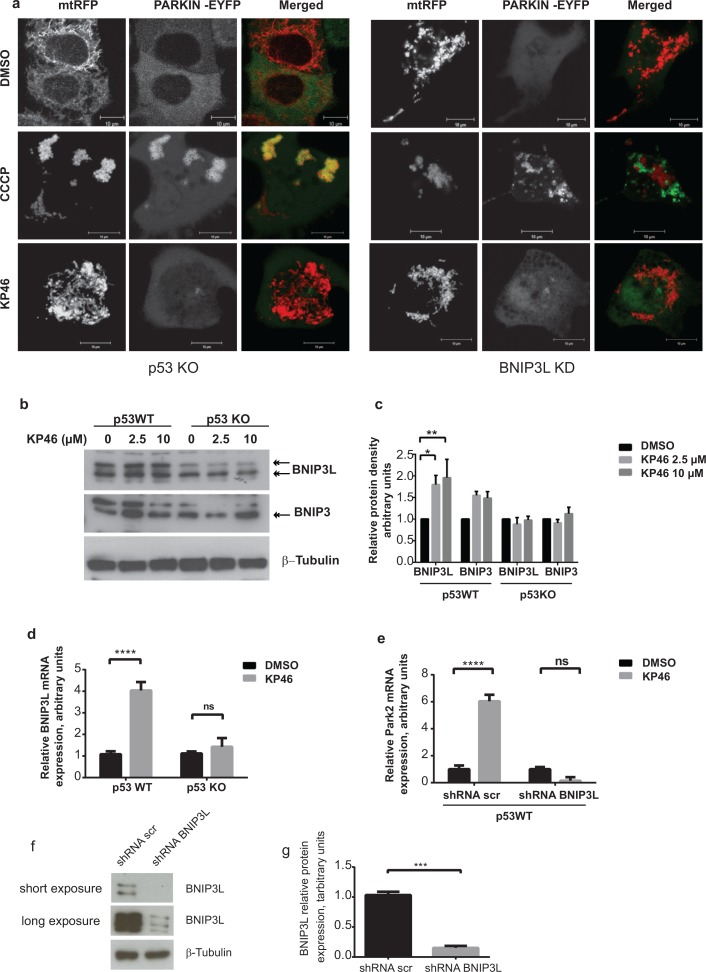
Figure 7. KP46 activates PARKIN in p53 and BNIP3L dependent manner.
a. HCT116p53KO and BNIP3LKD cells were co-transfected with PARKIN-EYFP (shown in green) and mtRFP (red) and exposed to vehicle or 2.5 μM KP46 for 4 hours or 50 μM CCCP as indicated respectively, for 2 hours and immediately monitored under confocal microscopy. Yellow overlay indicates the co-localisation of green and red fluorescence. Scale bars: 10 μm. b-c. KP46 upregulates BNIP3L in function of p53. b. Upregulation of BNIP3 and BNIP3L protein expression. HCT116WT or HCT116p53KO were exposed to vehicle or 2.5 or 10 μM KP46 for 6 hours. Protein lysates were immunoblotted with anti-BNIP3 and anti-BNIP3L antibodies or β-Tubulin (loading control). c. Quantification of Bnip3L and Bnip3 expression relative to β-Tubulin. n = 3 (n = 5 for Bnip3L in p53WT), ±SEM, two-way ANOVA, Bonferroni's multiple comparisons test. **p < 0.01, **p < 0.05 d. BNIP3L mRNA analyses of HCT116WT or HCT116 p53KO cells exposed to vehicle or 10 μM KP46 for 4 hours performed by RT-PCR. Shown is the mean normalized gene expression of BNIP3L ± SD (n = 3 individual experiments each in triplicates) two-way ANOVA followed by Bonferroni's multiple comparisons test, ****p < 0.0001 e. KP46 induces PARKIN in dependence of BNIP3L. PARK2 transcripts were analysed by RT-PCR of HCT116WT cells downregulated for BNIP3L (shRNA BNIP3L) or with scramble shRNA (shRNA scr) and exposed to vehicle (black bars) or 10 μM KP46 (grey bars) for 4 hours. Shown is the mean normalized gene expression of PARK2 ±SD (n = 3 independent measurements, carried out as triplicates). ****p < 0.001, two-way ANOVA, followed by Bonferroni's multiple comparisons test. f–g. Downregulation of BNIP3L. BNIP3L mRNA analyses of HCT116 expressing shRNA scramble or shRNABNIP3L cells exposed to vehicle or 10 μM KP46 for 4 hours performed by RT-PCR. Shown is the mean normalized gene expression of BNIP3L ±SD (n = 3 individual experiments each in triplicates), ***p = 0.0005, unpaired Students t-test, two-tailed (f) and the BNIP3L protein level in HCT116 expressing shRNA scramble or shRNABNIP3L (g) and the quantification of the interference, n = 3, ±SD, unpaired Students t-test, two-tailed, p = 0.0002.