Figure 3. RIP-Seq target validation.
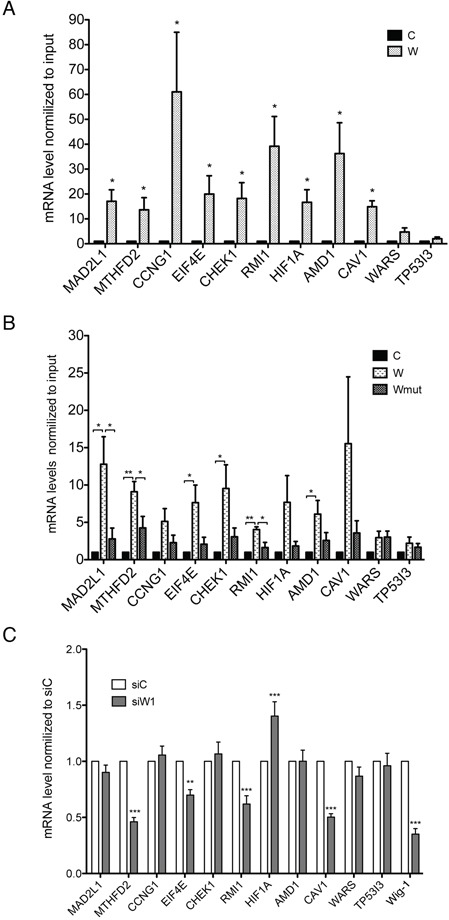
RIP was performed in HCT116 cells transiently transfected with Flag (C) or Flag-tagged Wig-1 (W) A. or in Saos-2 TetON cells (control, C) or Saos-2 TetON cells stably expressing either Flag-tagged wt Wig-1 (W) or a Flag-tagged zinc-finger 1 point mutant Wig-1 (Wmut) B. Wig-1 was precipitated with anti-Flag beads and the bound RNA was extracted. mRNA levels of the indicated targets were then determined by qRT-PCR and normalized to GAPDH mRNA and input. Data shown are mean ± SEM (n = 4 for HCT116 and n = 3 for Saos-2). *, p-value < 0.05 and **, p-value < 0.01 by Dunnett's multiple-comparison test. C. Wig-1 knockdown (siW1) leads to decreased levels of MTHFD2, EIF4E, RMI1 and CAV1 mRNAs and increased levels of HIF1A mRNA in HCT16 cells as assessed by qRT-PCR. Target mRNA levels were normalized to GAPDH mRNA, and control transfected cells (siC) are set to 1. Bars indicate mean ± SEM, n = 4. **, p-value < 0.01 and ***, p-value < 0.001 by Dunnett's multiple-comparison test.
