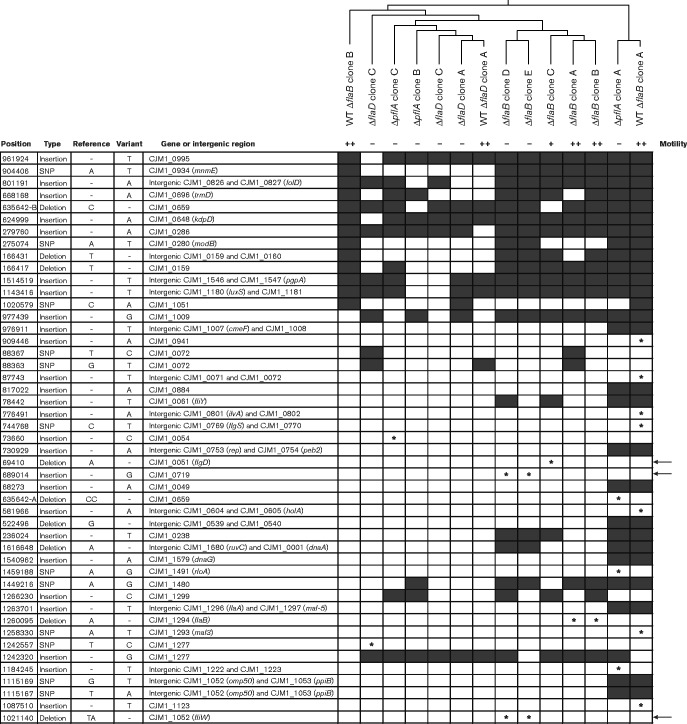
Fig. 3.
Genome sequencing revealed second-site mutations associated with motility defects. Genetic variation, i.e. SNPs and INDELs, in C. jejuni M1 defined gene deletion mutant clones and corresponding WT strains (Table 1) were analysed by Illumina sequencing; their presence is indicated with black boxes or with an asterisk when either found to be unique to individual mutant clones, WT strains, or associated with differential motility. Motility phenotypes are indicated as severely attenuated motility ( − ), intermediate motility (+) or WT motility (++) (see Fig. 1). SNPs or INDELs that are associated with attenuated motility in flaB (CJM1_1294) mutant clones C, D and E are indicated with an arrow on the right side of the panel. SNPs/INDELs that were detected in all sequenced M1 WTs and mutant strains compared to the M1 reference (Friis et al., 2010) were excluded from the analysis (Table S2). Hierarchical clustering was performed based on the presence/absence of SNPs and INDELs.