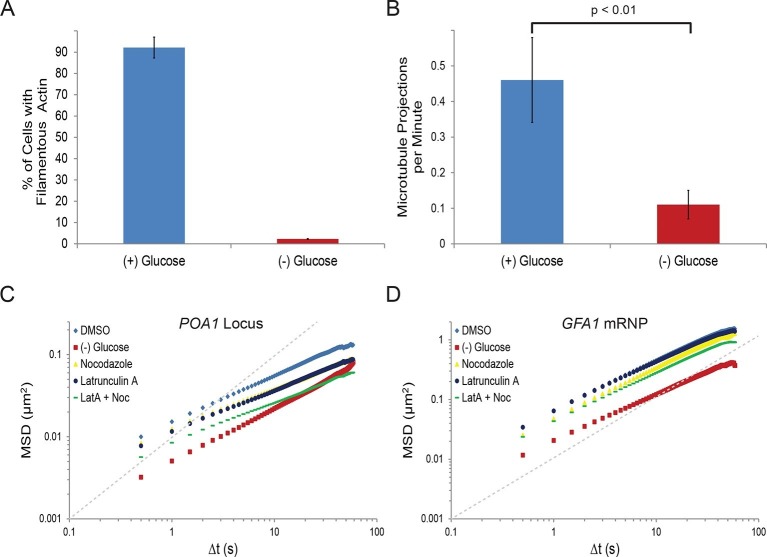
Figure 2. Starvation confines both cytoskeleton-independent macromolecular mobility and mobility influenced by the cytoskeleton (Figure 2—figure supplement 1).
(A) Quantification of filamentous actin during logarithmic growth, (+) glucose, and after acute starvation, (–) glucose. Cells were fixed and stained with phalloidin. Z-stack projections were then processed and cells were classified based on the presence or absence of filamentous actin. Error bars are standard error for three biological replicates (n = 406–709 cells per replicate). (B) Average number of microtubule projections per G1 cell during logarithmic growth, (+) glucose, and after acute starvation, (–) glucose. Error bars are standard deviation (SD) of the mean from three biological replicates (n = 10–1510-15 cells per replicate). The p-value for a two-tailed t-test for unpaired values assuming equal variance is shown. (C) Log-log MSD plot of the POA1 locus after treatment with nocodazole and/or latrunculin-A (LatA) for 20 min prior to imaging. (D) Log-log MSD plot of the GFA1 mRNP after treatment as described in (C). Dashed gray lines represent a slope of one to guide the eye.