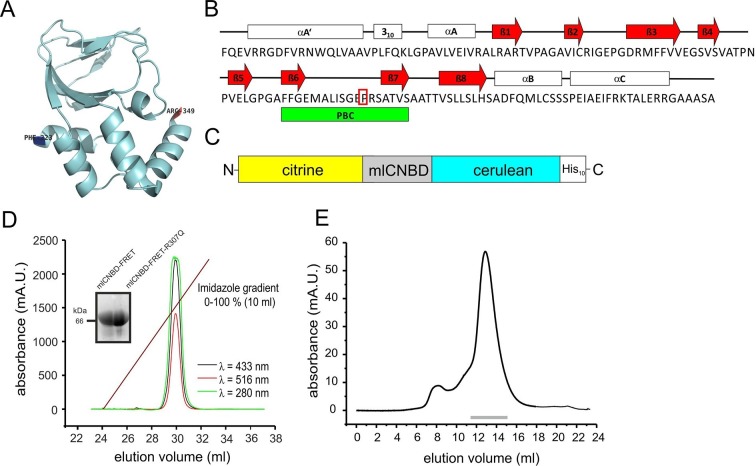
Figure 1. Generation and purification of mlCNBD-FRET.
(A) Ribbon presentation of a single mlCNBD according to (Schünke et al., 2011). The first (Phe223) and the last amino acid (Arg349) are indicated. (B) Structural features of mlCNBD. Alpha helices (αA-C), beta rolls (β1–8), and the phosphate binding-cassette (PBC) are indicated. The arginine (R) crucial for cAMP-binding is boxed. (C) The mlCNBD-FRET biosensor. The sensor has been generated by fusing citrine and cerulean to the N- and C-terminus of mlCNBD, respectively. For purification, a His10 tag has been added to the C-terminus. (D) Purification of mlCNBD-FRET via cobalt immobilized-metal affinity chromatography. Representative elution profile for mlCNBD-FRET using a linear imidazole gradient. The absorption has been recorded at three different wavelengths (280 nm: protein, green; 433 nm: cerulean, black; 516 nm: citrine, red). The inset shows a representative Western blot for the purified mlCNBD-FRET and mlCNBD-FRET-R307Q protein, stained with an anti-His antibody. (E) Size-exclusion chromatography of the purified mlCNBD-FRET protein. Representative elution profile. The protein eluted in a main peak at 67 kDa (peak maximum), close to the expected molecular mass of 70.9 kDa. A minor peak was observed that eluted earlier and represents the void volume. Fractions indicated by the grey line have been used for analysis.