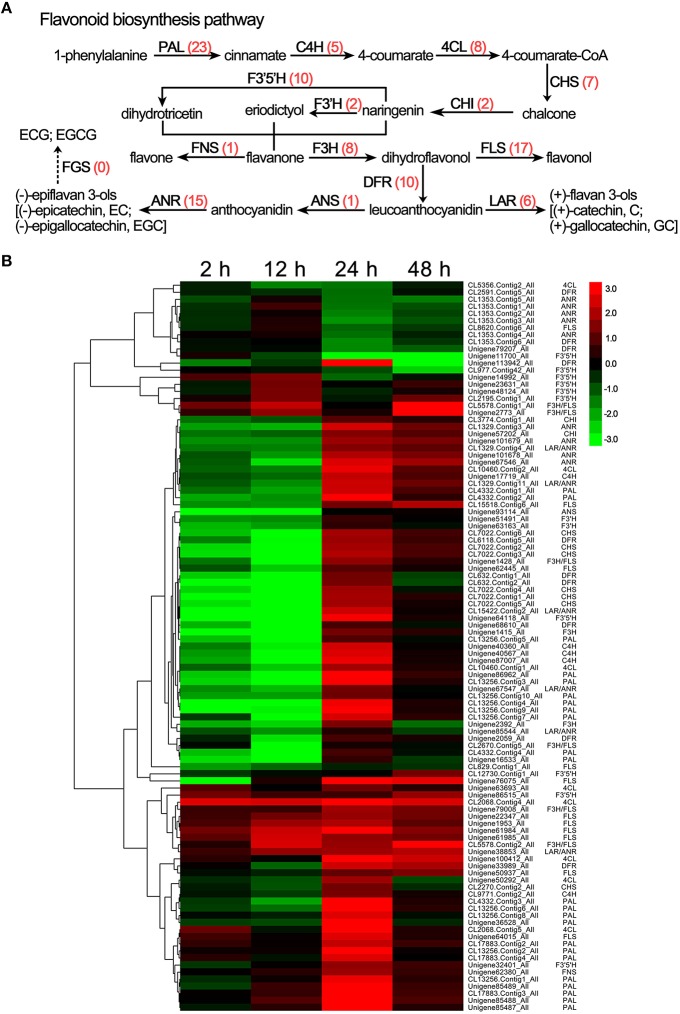
Figure 5.
DEGs involved in flavonoid biosynthesis in C. sinensis leaves under drought stress. (A) The flavonoid biosynthesis pathway. The red numbers in parentheses following each gene name indicate the number of corresponding DEGs. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; FNS, flavone synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; LAR, leucoanthocyanidin reductase; FGS, flavan-3-ol gallate synthase. (B) Hierarchical clustering analysis of the relative expression levels of the DEGs related to flavonoid biosynthesis. The FPKM ratio of unigene expression is represented on a logarithmic scale for each treatment period (2, 12, 24, or 48 h) and the control (0 h). Red indicates that a gene was up-regulated at that stage, whereas green indicates down-regulated expression.