Abstract
In vertebrates, neurotrophic signaling plays an important role in neuronal development, neural circuit formation, and neuronal plasticity, but its evolutionary origin remains obscure. We found and validated nucleotide sequences encoding putative neurotrophic ligands (neurotrophin, NT) and receptors (Trk and p75) in two annelids, Platynereis dumerilii (Errantia) and Capitella teleta (Sedentaria, for which some sequences were found recently by Wilson, 2009). Predicted protein sequences and structures of Platynereis neurotrophic molecules reveal a high degree of conservation with the vertebrate counterparts; some amino acids signatures present in the annelid Trk sequences are absent in the basal chordate amphioxus, reflecting secondary loss in the cephalochordate lineage. In addition, expression analysis of NT, Trk, and p75 during Platynereis development by whole-mount mRNA in situ hybridization supports a role of these molecules in nervous system and circuit development. These annelid data corroborate the hypothesis that the neurotrophic signaling and its involvement in shaping neural networks predate the protostome-deuterostome split and were present in bilaterian ancestors.
1. Introduction
During vertebrate development, differentiating neurons connect to themselves and to their target cells in order to generate a functional nervous system. Neurotrophic signaling ensures correct wiring, controlling cell survival and death, differentiation, neurite outgrowth, and target innervations [1–3] (as anticipated by the Nobel Prize Winners Levi-Montalcini and Cohen in 1986 [4, 5]). Neurotrophic signaling is also active in the adult nervous system, where it is involved in learning, memory, and plasticity, modulating long-term potentiation (LTP [6, 7]). Despite the importance of the neurotrophic signaling pathway for the functioning of the vertebrate nervous system, it is only very recently that its evolutionary origin and early function started to be revealed from invertebrate data [8–13]. In vertebrates, several neurotrophin ligands (NT), such as brain derived-neurotrophic factor (BDNF), nerve growth factor (NGF), NT3, and NT4/5 (and the NGF-related NT6/7 found in fish), bind to the high affinity tyrosine kinase receptors TrkA, TrkB, and TrkC, members of the RTK (receptor tyrosine kinase) superfamily, signaling through a tyrosine kinase intracellular domain (TK). They also bind to the low affinity coreceptor p75, member of the TNRF (tumor necrosis factor receptor) superfamily, signaling through an intracellular death domain (DD). Generally, upon neurotrophin binding, the Trk receptors are autophosphorylated in their TK domain and activate MAPK/ERK, AKT, and PLCγ signaling (promoting cell survival, cytoskeletal rearrangement, long-term potentiation, and neuronal plasticity in the growing neural circuits [3]). When the immature form of neurotrophin (proneurotrophin) binds p75 together with the Sortilin homodimer, it induces neuronal death and controls responses to neuronal injuries [14–16]. It is clear that much of the complexity of the neurotrophic signaling has evolved in the vertebrate lineage only, in the course of the two rounds of genome duplication. The jawless lampreys, for example, seem to possess only single ancestral forms of NT and two Trk receptors [17].
Neurotrophic signaling has long been considered a vertebrate novelty; yet, the cloning and characterization of neurotrophic signaling-related molecules also in invertebrates changed this view. A conserved Trk receptor was found in the cephalochordate amphioxus [10] and in other deuterostomes [18, 19]. Several neurotrophic signaling-related molecules were also found in various protostomes (such as Lymnaea [8, 9] and Drosophila [11]), suggesting an early bilaterian origin of neurotrophic signaling. This was recently confirmed via the identification of complete Trk, p75, and NT-like genes in the genome of the crustacean Daphnia pulex [12] and the isolation of a functionally equivalent Trk and neurotrophin molecule in mollusks [13]. Here, we investigate candidate ligands and receptors for neurotrophic signaling in Platynereis dumerilii, a marine annelid that belongs to the Errantia, a group of mostly freely moving annelid worms [20]. In comparison to other protostome models, Platynereis has undergone less evolutionary change yet is likewise amenable to molecular and genetic techniques and experimental manipulation; it is thus especially suitable for the study of ancestral molecules and cell types [21, 22]. Indeed, sequence comparison and prediction of domains and structure reveal the presence of canonical orthologs of NT, Trk, and p75 in Platynereis. Among known orthologs of nonchordate invertebrates, Platynereis neurotrophic molecules show the highest level of amino acid identity to the vertebrate counterparts found thus far. Further, whole-mount in situ hybridization analysis shows that these genes are expressed in the embryonic and larval central and peripheral nervous system. Furthermore, we identified a conserved NT ortholog in Capitella teleta, another annelid that belongs to the Sedentaria [20], an assembly of mostly sessile families. Therefore, together with recent findings in Ecdysozoa and other protostomes [12] and mollusks [13], our data support the notion that vertebrate-type NT, Trk, and p75 molecules existed in ancient annelids and bilaterians.
2. Materials and Methods
2.1. Platynereis dumerilii and Capitella teleta Culture
Platynereis dumerilii embryos and larvae were obtained from an established breeding culture at EMBL Heidelberg as described previously [21]. After fertilization, the embryos were raised in plastic cups, in natural seawater at 18°C, under standard light cycle conditions. Capitella teleta embryos and larvae were obtained as described [25].
2.2. Isolation of Platynereis and Capitella Neurotrophic Orthologs
Platynereis dumerilii (Table 1) sequence fragments were identified from available transcriptome and genome resources (Larsson et al., unpublished) upon BLAST searches with several domains of the vertebrate homologous sequences. A cDNA library was obtained from mixed larval stages between 24 hpf and 5 dpf using the GeneRacer Advanced RACE Kit (Life Technology) and the candidate sequence fragments were amplified and extended using standard polymerase chain reaction (PCR) and rapid amplification of cDNA ends (RACE). Capitella genes were retrieved from available resources (JGI genome portal from the Department of Energy Joint Genome Institute, University of California [26]) and compared to previously reported ones [12]. They were experimentally validated using a cDNA library of mixed stages. Genes were amplified using a high fidelity Phusion Polymerase (Thermo Fisher Scientific) (Table 1).
Table 1.
| Gene | Forward primer | Reverse primer |
|---|---|---|
|
Pdu Trk
(GenBank: KU206573) |
5′R_A: GGCACCTTTCCCAGACACAGGGCATCAGGGCC, 5′R_B: GCAGACTCCATAAAATGTCACAATATTCTCGTG, 5′R_C: CAAGGTACACTCTACCAAAAGCCCCTTCTCCTAATTC, 5′N_C: TCCTAATTCTCGAATAAATCTGATCTGCTT, 5′R_D: CAGGTAATTCGGATTATCCAACACATGAGG, 5′N_D: ACATGAGGTGCATTTAAGGGCATGGTC, 5′R_E: TCTTGGATCAACCTCAATTTCAGGGATTTGACA |
ATAAGAGACAGTAATCCGTAATTAAGCAATGA |
|
| ||
|
Pdu Nt
(GenBank: KU206574) |
GACGGAGGCTGGTCGCAAAAAACATGTCAC | GGGGGTATCACCGCATATCTTGCAGCAA |
|
| ||
|
Pdu p75
(GenBank: KU206572) |
5′R: AGTAATACCCCTGCCGACATTCGCAGACT, 5′N: GCAGACTGTGTCGTTAGTTATCGTACAGGG |
3′R: GCTTCATGTGTGCAACTACAGAGAGGATAC, 3′N: GATACAGTGTGTATGGAAATGCCCGTCCCAG |
|
| ||
| C.t. Trk | ATGTTTTTGAGTGACGTTGCGTGCT | ATCGGCGATGATTTCCAAATATGGTGG |
|
| ||
| C.t. Nt1 | ATGCAGCTTGATTGCTGGC (scaffold 4) | AGTCAGAGTTGCGGTACAGCA |
|
| ||
|
C.t. Nt2
(GenBank: KU206575) |
ATCGACATGCAGTGGAATCAAAGAAAATCC (scaffold669) | ATTTTCCATCAACTGAAATCGATCAGAC |
2.3. Domain Prediction and 3D In Silico Modeling
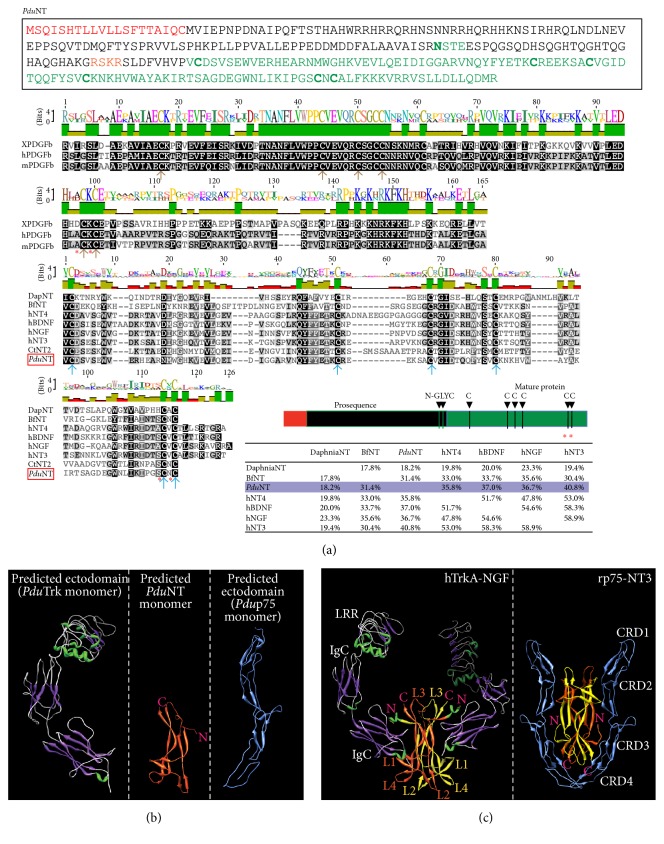
Protein domains were scanned and predicted using several tools from the Expasy suite: Prosite [27], SignalP 4.0 [28], and ProP 1.0 [29]. Protein 3D models were predicted using CPHmodels 3.2 [30] and M4T [31]. The predicted structures were visualized with Chimera (developed by the Resource for Biocomputing, Visualization, and Informatics, funded by the National Institutes of Health, NIGMS 9P41GM103311 [32]). For TrkA-NGF in Figure 2(c), the experimentally determined complexes in TrkA-NGF were used for comparison (DOI: 10.2210/pdb1www/pdb [23]). For p75-NT3 in Figure 2(c) the experimentally determined symmetrical complexes in p75-NT3 were used for comparison (DOI: 10.2210/pdb3buk/pdb [24]).
Figure 2.
Sequence analysis of PduNT and 3D modeling of the extracellular domain of PduTrk, NT, and p75. (a) Predicted a.a. sequence, schematic representation, and multiple sequence alignment for Platynereis NT. The alignment is done for the “Cys-Knot” domain (mature protein). In comparison the Cys-Knot of PDGF-beta is also shown. In the schematics in the lower right the different domains and the cysteines core (green) are indicated. Red labels the predicted signal peptide. N-glyc: putative glycosylation site. Species are indicated as in Figure 1. In the alignment, the cysteines forming the knot are shown with arrows (brown for the PDGF subfamily, blue for the NGF subfamily). (b) Predicted 3D structure of the extracellular domain of PduTrk (left panel), PduNT (middle panel), and Pdup75 (right panel). (c) As reference, a published 3D structure of the complex between the extracellular domain of TrkA-NGF (left panel, DOI: 10.2210/pdb1www/pdb [23]) and p75-NT3 (right panel, DOI: 10.2210/pdb3buk/pdb [24]) is shown. The domains are indicated (see text for details), as well as the N-terminus (N) and C-terminus (C) of the NTs (in pink), and the 4 loops formed by the NGF dimmer.
2.4. Phylogenetic Analysis
Sequence data were retrieved from Uniprot (The Universal Protein Resource [33]) and NCBI databases [34] or experimentally determined. For some genes we used the sequences indicated in Wilson, 2009 [12]. Sequence alignments were generated with Muscle [35] and visualized with Geneious [36]. Maximum likelihood (ML) trees were generated with PhyML [37], performing 100 bootstrap replicates with the LG substitution model.
2.5. Whole-Mount Single ISH Hybridization and Image Processing
Whole-mount in situ hybridization was performed as described [38], with slight modifications for detection of weakly expressed genes.
The full-length sequences or PduTrk, p75, and NT were cloned into PCR Topo II, and antisense RNA probes were transcribed in vitro. For staining of the nervous system a mouse antiacetylated tubulin (Sigma), at a dilution of 1 : 250, was used as primary antibody, while DyLight 488 anti-mouse (Jackson Laboratories) was used as secondary antibody, diluted 500 times. Images were processed with Fiji [39] or Imaris (Bitplane). Brightness and contrast were adjusted equally across the whole images. Images were cropped and processed in Illustrator to assemble figures.
3. Results and Discussion
3.1. Isolation and Characterization of Platynereis Trk, p75, and NT
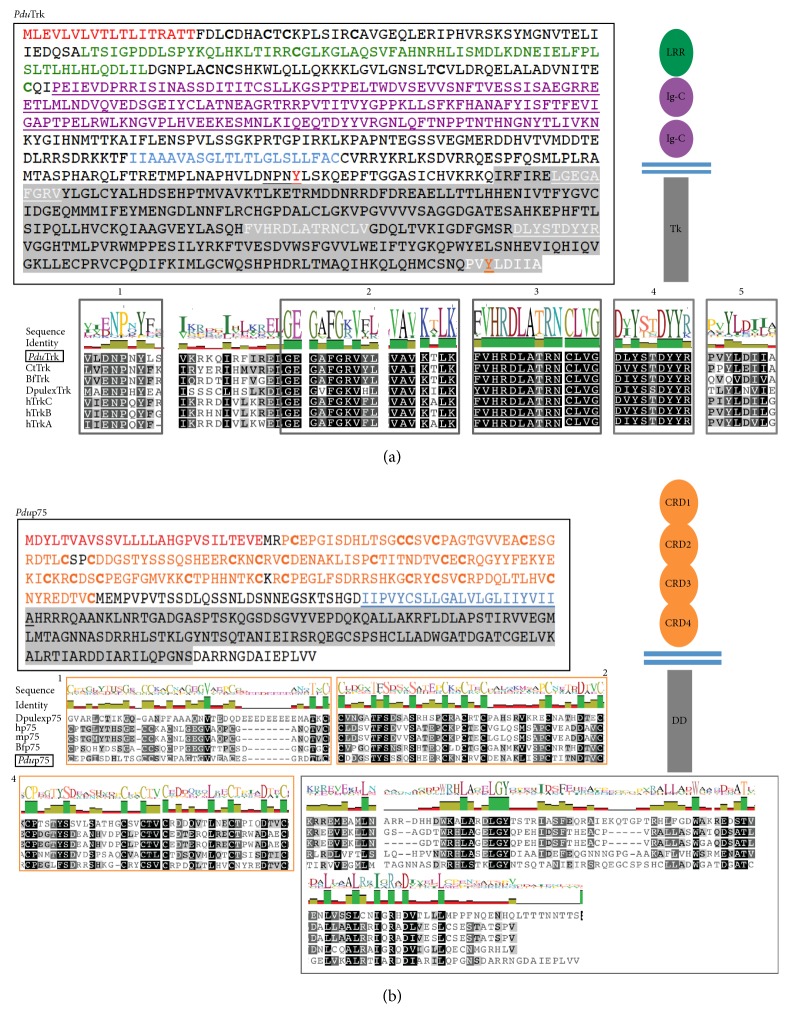
An intracellular tyrosine kinase (TK) domain was retrieved from a Platynereis EST collection with several features diagnostic of Trk-like receptors (Figure 1(a), gray in the amino acids sequence, and Figure S1, in Supplementary Material available online at http://dx.doi.org/10.1155/2016/2456062): a binding domain for PTB containing proteins (1), an ATP binding site (2), a catalytic domain with the key amino acid aspartate (3), and an autophosphorylation loop (4). We found that the Platynereis Trk TK domain even contains a conserved docking site for the PLCγ [vertebrates: P(VIS)YLD(IV)L(GE), Platynereis: PVYLDIIA] (5 in Figure 1(a) and Figure S1(c)) that, in vertebrates, catalyses the formation of DAG (Diacylglycerol) and IP3 (Inositol Triphosphate) from PIP2 (Phosphatidylinositol 4,5-Bisphosphate) upon activation and initiates a pathway implicated in cytoskeletal rearrangement, long-term potentiation, and neuronal plasticity [40, 41]. Notably, conservation of the tyrosine for the PLCγ docking site is also observed in the Trk ortholog of Daphnia pulex (DpulexTrk, although the amino acids surrounding the tyrosine do not seem to be conserved in Daphnia, Figure 1 and Figure S1 [12]) and in Aplysia [13]. Conversely, this docking site is completely absent in the basal chordate amphioxus [10, 12] (Figure 1(a), Figure S1), which indicates that the amphioxus Trk ortholog has been subject to secondary loss. To determine whether Platynereis Trk has a chordate-like N-terminal NT-binding domain, we extended the sequence by 5′ RACE and found highly stereotypical extracellular domains (similar to DpulexTrk, Figure 1(a), Figure S1). These comprised cell adhesion modules and characteristic clusters of cysteines (bold in Figure 1(a)). Beyond the signal peptide, two LRR (leucine rich repeats) domains are present (green in Figure 1(a)), followed by two predicted IgG (immunoglobulin) domains (purple in Figure 1(a)) that mediate NT binding in vertebrates [42]. The full-length version of Platynereis Trk shares 36,7% amino acid identity with the human TrkB, while its highly conserved TK domain shares more than 60% identity with the human TrkB (a high conservation, as compared to the Trk ortholog recently found in mollusks [13]). Thus, Platynereis Trk is the protostome Trk ortholog with the highest degree of amino acid identity to the vertebrate Trk counterpart. The evidences from Daphnia, Aplysia, and Platynereis indicate that the central core of the extracellular immunoglobulin domains was assembled before the protostome-deuterostome split, contrary to previous belief. It is plausible that Lymnaea lost one canonical IgG domain during evolution, due to domain reshuffling [43], but retained the ability to exert trophic functions [9].
Figure 1.
Sequence analysis of Platynereis Trk (PduTrk) and p75 (Pdup75) shows conserved domains. (a) Predicted a.a. sequence of Platynereis Trk, multiple sequence alignment for Platynereis Trk TK intracellular domain. Predicted domains are outlined in different colors in the sequence and represented in the schematic drawing on the right. LRR, green: leucine rich repeats domains, IgG, purple: immunoglobulin domain, light blue: transmembrane domain, and TK, gray: intracellular tyrosine kinase domain. Important signatures are also highlighted in the sequence, numbered in the alignment (1–5), and described more in detail in the text. 1: juxtamembrane domain, Src binding site, 2: ATP binding site, 3: conserved aspartate (D) in the catalytic site, 4: autophosphorylation domain, and 5: binding for PLCγ. (b) Predicted a.a. sequence and multiple sequence alignment for Platynereis p75. Predicted domains: CRD1–4, orange: cysteine rich domains, light blue: transmembrane domain, and DD, gray: intracellular death domain. The alignment shows the extracellular CRD1, CRD2, and CRD4 domains and the intracellular portion, containing also the DD domain. The alignment for the less conserved CRD3 is not shown. In both red labels there is the predicted signal peptide. Pdu: Platynereis dumerilii, Ct: Capitella teleta, Bf: Branchiostoma floridae, Dpulex: Daphnia pulex, h: Homo sapiens, and m: Mus musculus.
Next, we isolated and RACE-extended the sequence of Platynereis p75 (Figure 1(b), Figure S2), the putative ortholog of the vertebrate low affinity coreceptor p75 that binds to the complex of Trk/NT [2, 3]. Similar to Trk, Platynereis p75 exhibits several conserved features (Figure 1(b) and Figure S2). The intracellular part contains a canonical death domain (DD, in the gray region highlighted in the amino acids sequence), known to activate caspases implicated in cell death and common to all the death receptors of the TNFRSF superfamily (to which p75 belongs). This domain is absent from the TNFR found in Drosophila (Wengen [44]). However, because the death domain is present in p75 orthologs of arthropods and other protostomes ([12] and this study), it is likely that a receptor with such domain was established already at the base of Bilateria and lost or highly modified in some lineages. Accordingly, Drosophila also seems to have lost a fully assembled Trk gene or it is not possible to recognize due to the high level of molecule divergence, similar to the presence of multiple neurotrophin molecules with a highly divergent primary sequence [11]. Four prototypical CRDs (cysteine rich domains) constitute the extracellular portion of Platynereis p75 (orange in Figure 1(b)), similar to p75 in vertebrates and other deuterostomes. Notably, Daphnia p75 harbors 3 conserved CRDs only [12].
After a putative neurotrophic receptor (Trk) and coreceptor (p75) had been successfully characterized, we next set out to identify a putative candidate NT ligand. This search was more challenging as the vertebrate neurotrophins evolve relatively fast, concomitant with the extracellular domains of Trk and other RTK receptors [45]. Nevertheless, we could find one single hit for a possible NT-like molecule in the Platynereis transcriptome. Protein sequence analysis predicted that, similar to its vertebrate counterparts, Platynereis NT is composed of a signal peptide sequence comprising the first 20 amino acids and a proneurotrophin domain containing an N-glycosylation site, which is important for secretion (Figure 2(a)). Following the predicted protease cleavage site (RSKR) a core of around 120 amino acids rich in cysteines is found (Figure 2(a), green in the sequence overview). Amino acid sequence analysis and multiple sequence alignment (Figure 2(a)) show that this core of cysteines resembles the one present in the vertebrate neurotrophins (which fold into a Cys-Knot). As observed in the vertebrate neurotrophin sequences, the Cysteine “5” in the putative core of Platynereis NT is flanked by highly hydrophilic amino acids (serine and asparagine, red asterisks in Figure 2(a)), a key feature to predict the typical Cys-Knot foldings [46, 47]. A special motif of knotted cysteines folded by disulfide bonds is typical of growth factors and many extracellular proteins and hormones harbor such a motif (such as transforming growth factor-β, TGF-β, and platelet-derived growth factors, PDGF) [46, 48]. In these signaling molecules, such folding exposes the hydrophobic residues on the surface and helps the dimerization. Differences exist between the Cys-Knot of the different growth factor subfamilies, but all comprise 6 cysteines essential for the formation of disulfide bonds, which fold the special loops inside the β strands of these molecules [48], most likely an old molecular invention. The Cys-Knot of Platynereis NT belongs to the one shared by all neurotrophin molecules (Figure 2(a)). Further, as compared to the recently discovered Aplysia NT [13], we found that Platynereis NT shares a high amino acid identity with the vertebrate counterparts (Figure 2(a)). It shares more than 40% identity with human NT3 (in contrast to the 30% of Aplysia NT), 36,7% with human NGF (in contrast to the 32% of Aplysia NT), and 37% with BDNF (in contrast to the 28% of Aplysia NT). Thus, Platynereis neurotrophin is the most conserved protostome neurotrophin found so far.
In vertebrates, specific foldings of the Trk extracellular portion composed of IgG and LRR domains and of the p75 receptor (composed of 4 CRD) allow NT binding ([23, 24]). Thus, to further challenge sequence similarity and to assess whether Platynereis Trk, NT, and p75 might produce similar foldings to the vertebrate counterparts, we employed homology-based modeling algorithms and performed 3D structure prediction for Platynereis Trk, NT, and p75 (Figures 2(b) and 2(c)). These algorithms do not rely on a set protein or any a priori folding knowledge but identify and align related proteins, based on a PSI BLAST search and secondary structure prediction, upon which a 3D model is finally built. According to the homology-based 3D structure prediction, the Platynereis Trk extracellular domains are expected to assemble in canonical β-helices of the LRR and the IgG domains (Figure 2(b)), closely resembling the vertebrate situation (compare with the left panel of Figure 2(c) that shows experimentally obtained structures of human TrkA and NGF [23]).
Similarly, for Platynereis NT we predicted prototypical antiparallel β-sheets and heel-like folds that in vertebrates are determined by the Cys-Knot (Figure 2(b), middle panel). This special arrangement exposes the hydrophobic amino acids on the surface, which mediates homodimerization of neurotrophin monomers [49].
Similar to the vertebrate ortholog, our 3D modeling of the Platynereis p75 ectodomain predicts β-sheets folding into an elongated structure, with convex patches formed at the CRD2 site and between CRD3 and CRD4 (Figure 2(b), right panel); these sites have shown to be the binding patches for the neurotrophins dimmer (Figure 2(c), right panel) [24]. Our in silico data are only predictive and need to be validated experimentally. Biochemical analysis and crystal structures studies of the ectodomains binding to the ligands are needed to rule out whether this degree of amino acid conservation truly translates into functionally conserved domains. Nevertheless, as the models are based on primary and secondary structure alignments, these predictions are a strong indication that the extracellular domain of Platynereis Trk and p75 and the Platynereis NT possess all requisites to fold and bind to each other in a vertebrate-like fashion.
Amino acid sequence and structure prediction, as well as reciprocal BLAST, suggested that the single Platynereis Trk, p75, and NT proteins are orthologous to the vertebrate Trks, p75, and NTs. To confirm, we assessed the phylogenetic relationships of Platynereis neurotrophic signaling-related genes to their vertebrate and invertebrate counterparts.
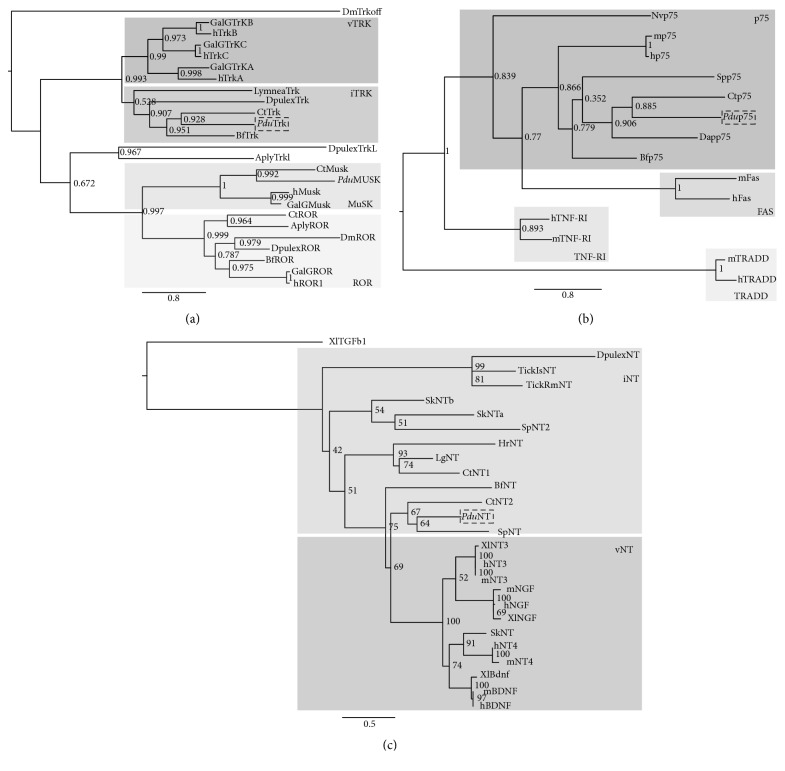
Maximum likelihood (ML) analysis supports orthology of Platynereis Trk (Figure 3(a)) that, as expected, clusters with other invertebrate Trk orthologs (“iTrk” in Figure 3(a)) and is most closely related to Capitella Trk. None of the invertebrate Trk sequences is more closely related to a specific vertebrate Trk type, suggesting that the latter are vertebrate-specific. Our tree likewise resolves specific clusters of Ror and Musk genes (including invertebrate members). Both Ecdysozoa (e.g., Daphnia) and Lophotrochozoa (e.g., Platynereis and Capitella) thus possess Trk, Ror, and Musk orthologs. These data support the hypothesis that the diversification of the RTK receptors into three different families (including true Trk receptors with their specific extracellular modules) predated the protostome-deuterostome split [45].
Figure 3.
Phylogenetic analysis of annelid Trk, p75, and NT. (a) Trk; (b) p75; (c) NT. Statistical support per node is shown. Outgroups or midbranching was used to root the trees; Platynereis sequences are indicated with dotted black squares. ML bootstrap values are indicated for 100 replicates. For comparison, different members of the RTK (a) and TNFRSF (b) superfamily are shown (details in the text). In (a) only the tyrosine kinase domain is used for the analysis, and in (c) only the mature protein (Cys-Knot) is used, i: invertebrate, and v: vertebrates. The species used for comparison are indicated as in Figures 1 and 2. In addition, Sp: Strongylocentrotus purpuratus, GalG: Gallus gallus, Lymnaea: Lymnaea stagnalis, Aply: Aplysia, Dm: Drosophila melanogaster, Hr: Helobdella robusta, and Nv: Nematostella vectensis. TickIs: Ixodes scapularis tick, TickRm: Rhipicephalus microplus tick, and Dap or Dpulex: Daphnia pulex.
The TNFRSF superfamily contains highly divergent members, separated into different family groups [50]. Our phylogenetic analysis resolves the separation of some of the TNFRSF into separate families and supports orthology of Platynereis p75 with vertebrate and invertebrate p75 proteins (Figure 3(b)). From this analysis it is also apparent that the divergence of these families most likely occurred before the divergence of deuterostomes and possibly was already established in the cnidarian-bilaterian ancestor (as suggested by the presence of a Nematostella p75, found performing BLAST searches against the Nematostella database: Nematostella vectensis v1.0 [51]).
Higher rates of evolutionary change and shorter overall sequence lengths render the phylogenetic analysis of neurotrophins more difficult. Nevertheless, ML analysis of the predicted mature core of Platynereis NT confirms orthology with vertebrate neurotrophin (Figure 3(c)). We could resolve the presence of two different branches (NT3-NGF and NT4-BDNF) that likely originated at the base of the vertebrates via duplication of a single ancestral neurotrophin [52]. Among the invertebrate sequences, our analysis resolves a group of ecdysozoan NTs (including Daphnia pulex NT), which is distinct from that of the Lophotrochozoa and from a cluster of deuterostome NTs (the acorn worm NTs: SkNTa and SkNTb and SpNT2) being closer to protostome rather than vertebrate homologs (confirming previous reports [12]). Further, we found and validated the presence of an additional NT in Capitella teleta (NT2, not reported previously). Despite the high degree of sequence divergence found in neurotrophin molecules we noted that, conversely to what has been previously reported based solely on CTN1, annelids seem to possess a more “chordate-like neurotrophin sequence” as Platynereis NT and the newly discovered Capitella NT (CTN2) fall into a group together with the chordate-like deuterostome NT of sea urchin (SpNT). Thus, it is likely that a prototypic neurotrophin already existed before the protostome-deuterostome divergence. Since the genomes of Platynereis and Strongylocentrotus may still be incomplete, we cannot exclude the presence of more protostome-like (in Platynereis) or chordate-like (in Strongylocentrotus) family members. In the absence of strong support for any of the basal bilaterian nodes, the exact number of neurotrophin paralogs necessarily remains unsettled. Similar to other extracellular ligands, neurotrophins may have duplicated several times in different lineages and generated highly divergent forms (such as the Spz in arthropods [11, 12] and the apCRNF in Aplysia [53]), which are difficult to recognize as putative neurotrophin orthologs [45]. Furthermore, the fast evolution and divergence of neurotrophin molecules might have triggered domain reshuffling of RTK receptors [43, 45] (possibly explaining the different forms of Trk-like molecules found in invertebrates, bearing losses and gain of various domains, such as for Trkl and Lymnaea Trk [12]). More exhaustive ortholog searches and phylogenetic analyses in additional protostome and deuterostome genomes will be needed to clarify the early stages of neurotrophin evolution.
3.2. Platynereis NT, Trk, and p75 Are Expressed in the Developing Embryonic and Larval Nervous System
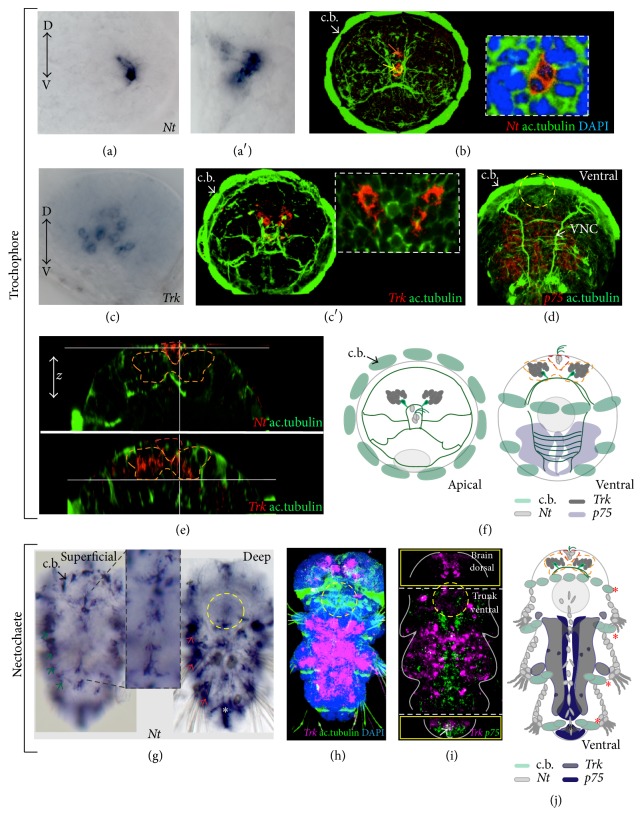
To gain insight into possible sites of activity of neurotrophic signaling in Platynereis, we investigated Nt, p75, and Trk mRNA expression at several larval stages. In the early trochophore larvae, Nt and Trk mRNA were detected in the brain (Figures 4(a)–4(c′), 4(e), and 4(f)) and p75 in the developing trunk nervous system (midline region and the left and right trunk, Figures 4(d) and 4(f)). We identified the cells producing NT (Figures 4(a)–4(b), 4(e), and 4(f)) as the apical tuft cells (yellow arrow in Figure 4(b)) and the crescent cells (orange arrow in Figure 4(b)) that are part of the apical organ. The apical organ differentiates first in the larval brain and is later complemented by developing adult brain parts [54]. Cells expressing Trk were found deeper in the dorsal developing adult brain (Figures 4(c), 4(c′), 4(e), and 4(f)), in cells that form part of a nonvisual, light sensitive region, which comprises vertebrate-type ciliary photoreceptors [21], produces melatonin, and harbors a circadian clock [55]. This early Nt and Trk expression suggests that the early forming apical organ emits neurotrophic signals that are received by cells of the later forming ciliary photoreceptor region and that Platynereis Trk might be involved in circadian clock entrainment (as reported for vertebrate TrkB [56]).
Figure 4.
Expression of PduNT, p75, and Trk in the developing worm. (a–b) Apical views. Expression of Nt in cells of the apical organ at 48 hpf ((a′) and the inset in (b) show a close-up on the Nt+ cells). Bright field images (a, a′), confocal z-projections (b). The yellow arrow indicates the apical tuft cells; the orange arrow indicates the crescent cells. (c, c′) Apical views. Expression of Trk in cells in the dorsal brain (the inset in (c′) shows a close-up on the Trk+ cells). Bright field image (c), confocal z-projection (c′). (d) Ventral view. Expression of p75 in the developing nervous system of the trunk, VNC: ventral nerve cord, confocal z-projection. In (a)–(d), D: dorsal, V: ventral. (e) Virtual cross sections show the position of the Nt+ (dashed red contour)/Trk+ cells (dashed orange contour) along the z-axis. (f) Schematic drawings of the apical view (left) and ventral view (right) showing Nt (light gray), Trk (dark gray), and p75 (light blue) expression of the trochophore larvae. (g) Superficial and deep Nt expression in the juvenile larva (nectochaete, around 3 dpf). Expression in the ciliary band (c.b.) along the superficial (green arrows) and deep (red arrows) periphery and in the midline (inset in (g)) is observed. (h) Trk expression in the ventral trunk nervous system and in the brain in the nectochaete stage. (i) Double WMISH showing the expression of Trk and p75 in the juvenile larva. The white arrow indicates colocalization at the posterior end. Different Z-projections from the same confocal scan are outlined by yellow squares: the upper one shows a single channel (magenta for Trk) within a z-projection of the dorsal brain (p75 mRNA is not found here); the lower square shows the posterior growth zone. (j) Schematic drawing of the expression of Nt, Trk, and p75 expression in the juvenile. In all the confocal images, the axonal scaffold of the nervous system is stained with acetylated tubulin (ac.tubulin, in green) and nuclei are blue (DAPI, 4′,6-diamidino-2-phenylindole). Asterisks: Nt expression in the pygidium (white in (g) and black in (j)); in (j) Nt expression in the ciliary bands (c.b., red asterisk) is indicated by a gray outline. A gray circle in (j) and a dashed yellow circle in the other panels indicate the stomodeum as reference.
At later larval stages (referred to as nectochaete [57]), when the trunk nervous system and musculature differentiate, Platynereis Nt expression was still observed in the apical organ region, but additional expression sites became apparent in the trunk. Those included the ciliary bands, the ventral midline, and peripheral superficial and deep cells (green and red arrows in Figure 4(g), schematics in Figure 4(j)), likely including also nonneuronal cells (such as prospective skin cells and developing muscle cells). At the same stages, Platynereis Trk is expressed broadly in the developing nervous system (Figures 4(h)–4(j)). Double WMISH revealed that while Trk and p75 clearly share a domain of expression at the posterior growth zone of the developing worm (white arrow in Figure 4(i)), their expression is almost mutually exclusive in the anterior nervous system. Here, Trk is expressed more broadly, at the left and right site of the neural tissue, while p75 is expressed mostly in cells surrounding the midline, where Nt is found (Figures 4(g)–4(j)). The more restricted expression of p75 around the site of NT secretion suggests that p75-NT might signal together independently from Trk, as reported from vertebrates. In Platynereis proliferating neuronal progenitors are found mostly on the surface of the ectoderm, in the medial-most domain around the wnt4+ midline (NT+), while differentiated neurons are located more laterally and basally [58]. It would be interesting to test whether NT secreted from Platynereis midline acts together with p75 in the nearby territory during neuronal progenitor proliferation, while Platynereis Trk signaling might be more predominant in differentiated neurons. For instance, in the vertebrate embryo target cells secrete neurotrophic ligands that attract the Trk+ axons of developing neurons, thus promoting innervation and also providing signals for neuronal survival [1]. Nt+ cells in Platynereis apical organ, neuronal midline, skin, and muscles might accordingly represent also a central and peripheral target sites for the outgrowing axons of developing Trk+ neurons, consistent with a role of the annelid neurotrophic signaling in axon pathfinding.
3.3. The Evolution of the Neurotrophic Signaling in Eumetazoa
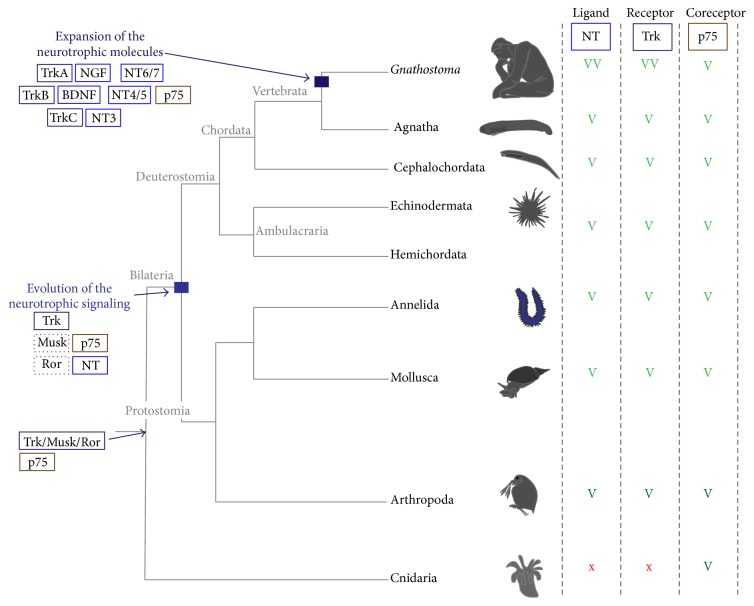
While it is clear that Trk, NT, and p75 were distinctly present and that the original tyrosine kinase had already split into different subfamilies in bilaterian ancestors (such as Trk, Ror, and Musk shown in Figure 3(a) [45]), the situation is less clear for bilaterian outgroups. According to Sossin [45], a common precursor molecule for Trk/Musk/Ror possessing a Frizzle/Kringle extracellular domain (similar to the ones present in Ror and Musk and in sponge tyrosine kinase receptors) and a conserved intracellular tyrosine kinase was likely present at the base of Eumetazoa. The split of the three families occurred only at the base of the Bilateria. In line with this, a Trk receptor and neurotrophin ligands appear to be absent from Nematostella ([59] and Antonella Lauri, unpublished), which seems to have only a p75 molecule (Figure 3(b)). Thus, a bona fide Trk and NT with their full complement of extracellular domains appear to have been evolved in the bilaterian stem line only (in contrast to p75, Figure 5).
Figure 5.
Evolution of the neurotrophic signaling predated the emergence of chordates. Eumetazoa tree showing the occurrence of NT, Trk, and p75 and the possible split of Trk from an ancestral Trk/Musk/Ror ancestor gene (after Sossin, 2006). A light green “V” is given when a conserved version of the protein is found; a dark green “V” indicates the presence of less conserved molecules (see text for details). Double light green “VV” indicates expansion of the molecules due to genome duplication. A red “x” is given when a putative ortholog is not found. Evolution of the neurotrophic signaling is hypothesized to have occurred at the base of Bilateria, while expansion of the single molecules likely occurred only in the vertebrate lineage.
4. Conclusions
In conclusion, our data on the highly conserved annelid neurotrophic molecules corroborate recent hypotheses that vertebrate-like representatives of neurotrophic signaling molecules existed at the base of Bilateria, predating the protostome-deuterostome split. The identification of a p75 ortholog in Nematostella furthermore indicates that this receptor was already present in the cnidarian-bilaterian ancestor. Urbilaterian neurotrophic signaling thus included a Trk receptor with a typical extracellular domain, as found in some extant, slow-evolving invertebrates (such as Platynereis) and in vertebrates, a p75 coreceptor, and one or more NT ligands, possibly performing trophic functions in the nervous system.
Supplementary Material
The Supplementary material provides detailed information on the full-length sequence of Platynereis Trk and p75, and on the amino acid identity as compared to other species.
Acknowledgments
The authors thank Dr. Tomas Larsson and Dr. Èlia Benito-Gutiérrez for discussion and inputs on the project, Dr. Tomas Larsson and Dr. Oleg Simakov for sharing the available transcriptome and genome dataset, Dr. Heather Marlow for the Capitella teleta cDNA library, Dr. Maria Antonietta Tosches for discussion on the gene expression, and Dr. Tomas Larsson for providing valuable input during the revision process. Antonella Lauri, Paola Bertucci, and Detlev Arendt acknowledge support by the European Molecular Biology Laboratory (EMBL). Antonella Lauri acknowledges support by the EMBL International Ph.D. Programme and by the European Union's Seventh Framework Programme project “Evolution of Gene Regulatory Networks in Animal Development (EVONET)” [215781-2]. Paola Bertucci and Detlev Arendt acknowledge support by a European Research Council grant (EA-AdG-2011-294810_BrainEvoDevo).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Bibel M., Barde Y.-A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes and Development. 2000;14(23):2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- 2.Huang E. J., Reichardt L. F. Neurotrophins: roles in neuronal development and function. Annual Review of Neuroscience. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang E. J., Reichardt L. F. Trk receptors: roles in neuronal signal transduction. Annual Review of Biochemistry. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S., Levi-Montalcini R., Hamburger V. A nerve growth-stimulating factor isolated from sarcomas 37 and 180. Proceedings of the National Academy of Sciences of the United States of America. 1954;40(10):1014–1018. doi: 10.1073/pnas.40.10.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levi-Montalcini R. Tissue and nerve growth promoting factors. Proceedings of the Royal Society of Medicine. 1965;58(5):357–360. [PMC free article] [PubMed] [Google Scholar]
- 6.Minichiello L., Korte M., Wolfer D., et al. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24(2):401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 7.Minichiello L., Calella A. M., Medina D. L., Bonhoeffer T., Klein R., Korte M. Mechanism of TrkB-mediated hippocampal long-term potentiation. Neuron. 2002;36(1):121–137. doi: 10.1016/S0896-6273(02)00942-X. [DOI] [PubMed] [Google Scholar]
- 8.van Kesteren R. E., Fainzilber M., Hauser G., et al. Early evolutionary origin of the neurotrophin receptor family. The EMBO Journal. 1998;17(9):2534–2542. doi: 10.1093/emboj/17.9.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck G., Munno D. W., Levy Z., et al. Neurotrophic activities of trk receptors conserved over 600 million years of evolution. Journal of Neurobiology. 2004;60(1):12–20. doi: 10.1002/neu.10329. [DOI] [PubMed] [Google Scholar]
- 10.Benito-Gutiérrez È., Nake C., Llovera M., Comella J. X., Garcia-Fernàndez J. The single AmphiTrk receptor highlights increased complexity of neurotrophin signalling in vertebrates and suggests an early role in developing sensory neuroepidermal cells. Development. 2005;132(9):2191–2202. doi: 10.1242/dev.01803. [DOI] [PubMed] [Google Scholar]
- 11.Zhu B., Pennack J. A., McQuilton P., et al. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biology. 2008;6(11, article e284) doi: 10.1371/journal.pbio.0060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson K. H. The genome sequence of the protostome Daphnia pulex encodes respective orthologues of a neurotrophin, a Trk and a p75NTR: evolution of neurotrophin signaling components and related proteins in the bilateria. BMC Evolutionary Biology. 2009;9, article 243 doi: 10.1186/1471-2148-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassabov S. R., Choi Y.-B., Karl K. A., Vishwasrao H. D., Bailey C. H., Kandel E. R. A single Aplysia neurotrophin mediates synaptic facilitation via differentially processed isoforms. Cell Reports. 2013;3(4):1213–1227. doi: 10.1016/j.celrep.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronfman F. C., Fainzilber M. Multi-tasking by the p75 neurotrophin receptor: sortilin things out? EMBO Reports. 2004;5(9):867–871. doi: 10.1038/sj.embor.7400219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnett M. G., Ryals J. M., Wright D. E. pro-NGF, sortilin, and p75NTR: potential mediators of injury-induced apoptosis in the mouse dorsal root ganglion. Brain Research. 2007;1183(1):32–42. doi: 10.1016/j.brainres.2007.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volosin M., Trotter C., Cragnolini A., et al. Induction of proneurotrophins and activation of p75NTR-mediated apoptosis via neurotrophin receptor-interacting factor in hippocampal neurons after seizures. The Journal of Neuroscience. 2008;28(39):9870–9879. doi: 10.1523/jneurosci.2841-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallböök F., Lundin L.-G., Kullander K. Lampetra fluviatilis neurotrophin homolog, descendant of a neurotrophin ancestor, discloses the early molecular evolution of neurotrophins in the vertebrate subphylum. The Journal of Neuroscience. 1998;18(21):8700–8711. doi: 10.1523/JNEUROSCI.18-21-08700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapraz F., Röttinger E., Duboc V., et al. RTK and TGF-β signaling pathways genes in the sea urchin genome. Developmental Biology. 2006;300(1):132–152. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Bothwell M. Evolution of the neurotrophin signaling system in invertebrates. Brain, Behavior and Evolution. 2006;68(3):124–132. doi: 10.1159/000094082. [DOI] [PubMed] [Google Scholar]
- 20.Struck T. H., Paul C., Hill N., et al. Phylogenomic analyses unravel annelid evolution. Nature. 2011;471(7336):95–98. doi: 10.1038/nature09864. [DOI] [PubMed] [Google Scholar]
- 21.Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodf J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306(5697):869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- 22.Lauri A., Brunet T., Handberg-Thorsager M., et al. Development of the Annelid Axochord: insights into notochord evolution. Science. 2014;345(6202):1365–1368. doi: 10.1126/science.1253396. [DOI] [PubMed] [Google Scholar]
- 23.Wlesmann C., Ultsch M. H., Bass S. H., de Vos A. M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401(6749):184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 24.Gong Y., Cao P., Yu H.-J., Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454(7205):789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 25.Seaver E. C., Thamm K., Hill S. D. Growth patterns during segmentation in the two polychaete annelids, Capitella sp. I and Hydroides elegans: comparisons at distinct life history stages. Evolution and Development. 2005;7(4):312–326. doi: 10.1111/j.1525-142x.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 26.Nordberg H., Cantor M., Dusheyko S., et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Research. 2014;42(1):D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigrist C. J. A., Cerutti L., de Castro E., et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Research. 2009;38(1):D161–D166. doi: 10.1093/nar/gkp885.gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen T. N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 29.Duckert P., Brunak S., Blom N. Prediction of proprotein convertase cleavage sites. Protein Engineering, Design and Selection. 2004;17(1):107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen M., Lundegaard C., Lund O., Petersen T. N. CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Research. 2010;38(supplement 2):W576–W581. doi: 10.1093/nar/gkq535.gkq535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Fuentes N., Madrid-Aliste C. J., Rai B. K., Fajardo J. E., Fiser A. M4T: a comparative protein structure modeling server. Nucleic Acids Research. 2007;35(2):W363–W368. doi: 10.1093/nar/gkm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen E. F., Goddard T. D., Huang C. C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEntyre J., Ostell J. The NCBI Handbook. Bethesda, Md, USA: National Center for Biotechnology Information; 2002. [Google Scholar]
- 35.Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearse M., Moir R., Wilson A., et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S., Lethiec F., Duroux P., Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research. 2005;33(2):W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tessmar-Raible K., Steinmetz P. R. H., Snyman H., Hassel M., Arendt D. Fluorescent two-color whole mount in situ hybridization in Platynereis dumerilii (Polychaeta, Annelida), an emerging marine molecular model for evolution and development. BioTechniques. 2005;39(4):460–462. doi: 10.2144/000112023. [DOI] [PubMed] [Google Scholar]
- 39.Schindelin J., Arganda-Carreras I., Frise E., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipsky R. H., Marini A. M. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Annals of the New York Academy of Sciences. 2007;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- 41.Minichiello L. TrkB signalling pathways in LTP and learning. Nature Reviews Neuroscience. 2009;10(12):850–860. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- 42.Urfer R., Tsoulfas P., O'Connell L., Shelton D. L., Parada L. F., Presta L. G. An immunoglobulin-like domain determines the specificity of neurotrophin receptors. The EMBO Journal. 1995;14(12):2795–2805. doi: 10.1002/j.1460-2075.1995.tb07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benito-Gutiérrez È., Garcia-Fernàndez J., Comella J. X. Origin and evolution of the Trk family of neurotrophic receptors. Molecular and Cellular Neuroscience. 2006;31(2):179–192. doi: 10.1016/j.mcn.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Kanda H., Igaki T., Kanuka H., Yagi T., Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. The Journal of Biological Chemistry. 2002;277(32):28372–28375. doi: 10.1074/jbc.c200324200. [DOI] [PubMed] [Google Scholar]
- 45.Sossin W. S. Tracing the evolution and function of the Trk superfamily of receptor tyrosine kinases. Brain, Behavior and Evolution. 2006;68(3):145–156. doi: 10.1159/000094084. [DOI] [PubMed] [Google Scholar]
- 46.Sun P. D., Davies D. R. The cystine-knot growth-factor superfamily. Annual Review of Biophysics and Biomolecular Structure. 1995;24(1):269–291. doi: 10.1146/annurev.bb.24.060195.001413. [DOI] [PubMed] [Google Scholar]
- 47.Vitt U. A., Hsu S. Y., Hsueh A. J. W. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Molecular Endocrinology. 2001;15(5):681–694. doi: 10.1210/me.15.5.681. [DOI] [PubMed] [Google Scholar]
- 48.Murray-Rust J., McDonald N. Q., Blundell T. L., et al. Topological similarities in TGF-β 2, PDGF-BB and NGF define a superfamily of polypeptide growth factors. Structure. 1993;1(2):153–159. doi: 10.1016/0969-2126(93)90029-g. [DOI] [PubMed] [Google Scholar]
- 49.Butte M. J. Neurotrophic factor structures reveal clues to evolution, binding, specificity, and receptor activation. Cellular and Molecular Life Sciences. 2001;58(8):1003–1013. doi: 10.1007/pl00000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bridgham J. T., Wilder J. A., Hollocher H., Johnson A. L. All in the family: evolutionary and functional relationships among death receptors. Cell Death and Differentiation. 2003;10(1):19–25. doi: 10.1038/sj.cdd.4401174. [DOI] [PubMed] [Google Scholar]
- 51.Putnam N. H., Srivastava M., Hellsten U., et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 52.Hallböök F. Evolution of the vertebrate neurotrophin and Trk receptor gene families. Current Opinion in Neurobiology. 1999;9(5):616–621. doi: 10.1016/S0959-4388(99)00011-2. [DOI] [PubMed] [Google Scholar]
- 53.Pu L., Kopec A. M., Boyle H. D., Carew T. J. A novel cysteine-rich neurotrophic factor in Aplysia facilitates growth, MAPK activation, and long-term synaptic facilitation. Learning and Memory. 2014;21(4):215–222. doi: 10.1101/lm.033662.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marlow H., Tosches M. A., Tomer R., et al. Larval body patterning and apical organs are conserved in animal evolution. BMC Biology. 2014;12, article 7 doi: 10.1186/1741-7007-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosches M. A., Bucher D., Vopalensky P., Arendt D. Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell. 2014;159(1):46–57. doi: 10.1016/j.cell.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang F.-Q., Walline R., Earnest D. J. Circadian rhythm of brain-derived neurotrophic factor in the rat suprachiasmatic nucleus. Neuroscience Letters. 1998;242(2):89–92. doi: 10.1016/S0304-3940(98)00062-7. [DOI] [PubMed] [Google Scholar]
- 57.Fischer A. H. L., Henrich T., Arendt D. The normal development of Platynereis dumerilii (Nereididae, Annelida) Frontiers in Zoology. 2010;7, article 31 doi: 10.1186/1742-9994-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demilly A., Steinmetz P., Gazave E., Marchand L., Vervoort M. Involvement of the Wnt/β-catenin pathway in neurectoderm architecture in Platynereis dumerilii . Nature Communications. 2013;4, article 1915 doi: 10.1038/ncomms2915. [DOI] [PubMed] [Google Scholar]
- 59.Lauri A. The evolution of the neural crest from an annelid perspective: conserved cell types and signaling pathways in Platynereis dumerilii [Ph.D. thesis] Heidelberg, Germany: Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary material provides detailed information on the full-length sequence of Platynereis Trk and p75, and on the amino acid identity as compared to other species.