Abstract
The E-cadherin gene (CDH1) is associated with poor prognosis and metastasis in patients with breast cancer, and methylation of its promoter is correlated with decreased gene expression. However, there is currently no direct evidence that CDH1 promoter methylation indicates poor prognosis in patients with breast cancer. In the present study, methylation-specific polymerase chain reaction (PCR) was applied to detect the methylation status of the CDH1 promoter in 137 primary breast cancer, 85 matched normal breast tissue and 13 lung metastasis specimens. Reverse transcription-quantitative PCR was used to assess the relative expression levels of CDH1 mRNA, and correlation analysis between CDH1 methylation status, and gene expression, clinicopathological characteristics and patient survival was performed. Methylation of CDH1 was identified in 40.9% (56/137) of primary breast cancer specimens, 61.5% (8/13) of lung metastasis specimens and none of the matched normal breast specimens. The downregulation of CDH1 mRNA and E-cadherin protein expression were identified to be significantly correlated with CDH1 methylation (P<0.05). In addition, CDH1 methylation was significantly associated with lymph node metastasis and estrogen receptor status of patients (P<0.05). In univariate analyses, patients with CDH1 methylation exhibited poor overall survival (OS) and disease-free survival (DFS; P<0.05). Furthermore, multivariate analyses revealed that CDH1 methylation was an independent prognostic factor predicting poor OS (HR, 1.737; 95% CI, 0.957–3.766; P=0.041) and DFS (HR, 2.018; 95% CI, 2.057–3.845; P=0.033) in patients with breast cancer. Therefore, the present study suggests that CDH1 promoter methylation may be correlated with breast carcinogenesis and indicates poor prognosis in patients with breast cancer.
Keywords: breast cancer, E-cadherin gene, methylation, lymph node metastasis, prognosis
Introduction
Breast cancer is the most frequently diagnosed type of cancer and the leading cause of cancer mortality among women (1). Despite the recent trend in decreasing in mortality rates due to the improvements in early detection and treatment, 458,400 mortalities were attributed to breast cancer in 2008 (1).
The development of breast cancer is generally considered to be a result of complex genetic and epigenetic alternations (2,3). Although cancer initiation and progression are predominantly driven by acquired genetic alterations, it is becoming clear that microenvironment-mediated epigenetic perturbations have an important role in neoplastic development. A number of well-characterized epigenetic modifications, including the DNA methylation of Ras association domain family member 1 (RASSF1), estrogen receptor 1 (ER), progesterone receptor (PR), retinoic acid receptor β, cyclin D2 and paired like homeodomain 2, and the acetylation and methylation of histones, are associated with aberrant gene functions and altered patterns of gene expression that are critical in breast cancer (4). In addition, secretoglobin family 3A member 1, O-6-methylguanine-DNA methyltransferase and RASSF1 promoter methylation had been previously considered as suitable biomarkers for the detection of field cancerization in breast cancer (5).
E-cadherin protein is encoded by the E-cadherin gene (CDH1; 16q22.1) and is the prototypical type I cadherin, a transmembrane glycoprotein mediating hemophilic cell-cell adhesion between neighboring cells (6). The E-cadherin-catenin complex, comprised of intracellular domains of E-cadherin and catenin, can activate certain signaling cascades and has an active role in epithelial-mesenchymal transition (EMT) (7,8). During tumorigenesis, E-cadherin has an important role in suppressing invasion and metastasis of breast cancer cells. As a result, decreased expression of E-cadherin is associated with an increased aggressive behavior in clinical breast cancer (9).
Complete loss of E-cadherin protein expression was identified in 84% of lobular breast carcinomas (10). Mutational inactivation of CDH1 accompanied by loss of heterozygosity (LOH) of the wild-type allele was detected in 56% of lobular breast carcinomas (11). Furthermore, gene knock-out experiments in mice demonstrated that CDH1 mutations were causal for the lobular breast cancer phenotype (12). Thus, invasive lobular breast carcinomas are characterized by the complete loss of E-cadherin expression caused by inactivating mutations and deletions.
It has also been reported that E-cadherin protein expression is reduced or absent in breast invasive ductal carcinoma; however, CDH1 mutations were rare or absent (13). It is of note that CDH1 promoter methylation is another important mechanism for inhibition of E-cadherin protein expression. This mechanism has been confirmed in breast cancer cell lines, however, data derived from primary breast cancer tissues is limited (14,15). Therefore, more research is required to precisely identify the correlation between CDH1 methylation and the prognosis of patients with breast cancer.
In the present study, the methylation status and mRNA expression level of CDH1 were detected in breast cancer tissues and the matched normal breast tissues. In addition, the correlation of CDH1 promoter region methylation was analyzed with the characteristics and prognosis of patients with breast cancer.
Materials and methods
Patients
A total of 245 specimens (137 primary breast cancers, 85 matched normal breast specimens, 13 lung metastasis specimens and 10 normal breast tissues corresponding to benign lesions) were obtained from 160 patients at the First Affiliated Hospital of Xi'an Jiaotong University (Xi'ian, China) between March 2007 and October 2009. The specimens were resected and frozen in liquid nitrogen immediately after surgery. This study was approved by the Ethics Committee of Xi'an Jiaotong University First Affiliated Hospital and each participant signed an informed consent document. The patients were accrued consecutively and the inclusion criteria were no previous histological diagnosis of breast cancer. All patients had undergone segmental resection or mastectomy and none had received radiotherapy or chemotherapy prior to surgery. The evidence of cancer, including the lymph node metastasis, tumor size and status of ER, PR, human epidermal growth factor receptor-2 (Her-2), tumor protein 53 (p53), Ki-67 and E-cadherin, was based on documented medical records. The clinical tumor node metastasis (TNM) stage was determined according to American Joint Committee on Cancer staging manual (16). Histological grading was made according to the Elston-Ellis modification of Scarff-Bloom-Richardson grading system (17). The 137 patients with primary breast cancer were followed up for 72 months to determine overall survival (OS) and disease-free survival (DFS).
Reverse transcription-quantitative polymerase chain reaction (PCR)
Following mechanical tissue homogenization, total RNA was isolated from the fresh clinical specimens using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacture's protocol. Concentration and purity of total RNA were assessed using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Approximately 1 µg of total RNA from each sample was reverse-transcribed into single strand cDNA (final volume, 10 µl) using PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol.
The following primers were used for amplification: Forward, 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′ for GAPDH (140-bp fragment); and forward, 5′-TGCTCTTGCTGTTTCTTCGG-3′ and reverse, 5′-TGCCCCATTCGTTCAAGTAG-3′ for CDH1 (423-bp fragment). The reaction was performed using Takara Taq DNA Polymerase Hot Start Version (Takara Biotechnology Co., Ltd.) in a total volume of 25 µl, containing 0.125 µl Takara Taq HS (5 U/µl), 0.25 µl of each pair of primers (20 µM), cDNA template 2 µl, 2 µl dNTP Mixture (2.5 mM each) and 2.5 µl 10X PCR Buffer. The amplification was conducted by performing 35 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec and extension at 72°C for 1 min in a PTC-200 thermal cycler (Bio-Rad Laboratories), according to the manufacturer's protocol. The post-amplification specific PCR products were separated on a 2.5% Biowest Regular Agarose G-10 (Gene Company Ltd., Chi Wan, Hong Kong) and analyzed using electrophoresis.
The primers for target gene (CDH1) and internal control gene [β-actin (ACTB)] were as follows: Forward, 5′-CAGCACGTACACAGCCCTAA-3′ and reverse, 5′-ACCTGAGGCTTTGGATTCCT-3′ for CDH1; and forward, 5′-TTCTACAATGAGCTGCGTGTG-3′ and reverse, 5′-GGGGTGTTGAAGGTCTCAAA-3′ for ACTB. qPCR was conducted in a total reaction volume of 25 µl, containing 2 µl cDNA, 12.5 µl SYBR Premix Ex Taq II (Tli RNaseH Plus, 2X; Takara Biotechnology Co., Ltd.), 8.5 µl dH2O and 1.0 µl of each pair of primers (10 µM). The amplification was performed as follows: Pre-denaturation at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, according to the manufacturer's protocol. The quantification cycle (Cq) was automatically calculated using iQ5 Optical System Software version 2.1 (Bio-Rad Laboratories, Hercules, CA, USA). Relative expression levels of the target gene were determined using internal control (ACTB). Data were analyzed using the quantitative threshold cycle (2−ΔΔCq) method (18). All amplification reactions were performed in triplicate. The CDH1 mRNA expression level was detected by qPCR, and the change fold relative expression level was calculated as a ratio of the average expression level of normal control tissues, from patients with benign lesions.
DNA extraction and treatment with sodium bisulfite
Genomic DNA was isolated from tissues using a TIANamp Genomic DNA kit (Tiangen, Beijing, China), according to the manufacturer's protocol. DNA concentrations and purity were measured by a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.).
The conversion of DNA by sodium bisulfite was performed according to the following established protocol. Initially, 2 µg of genomic DNA were denatured with 3 M NaOH at 42°C for 30 min (final concentration, 0.3 M NaOH), followed by incubation with freshly prepared 2.5 M sodium bisulfite (cat no. S9000) and 1 M hydroquinone (pH 5.0; cat no. H9003; Sigma-Aldrich, St. Louis, MO, USA) to a total volume of 520 µl at 55°C for 16 h. The DNA was purified with the Wizard DNA Clean-Up System (Promega, Madison, WI, USA), according to the manufacturer's protocol. Modification of the DNA was terminated by the addition of 5.5 µl NaOH (3 M) at room temperature for 15 min. The precipitation was conducted through the addition of 33 µl ammonium acetate (10 M, pH 7.0), 270 µl ethanol and 4.0 µl glycogen (10 µg/µl; Invitrogen; Thermo Fisher Scientific, Inc.), at −20°C for 12 h. The modified DNA was resuspended in 30 µl elution buffer and stored at −20°C.
Methylation-specific-PCR (MS-PCR)
The sodium bisulfite-converted DNA was amplified from 50 ng with Takara Taq Hot Start Version (Takara Biotechnology Co., Ltd.), using the following protocols. The polymerase chain reaction amplification was performed using Takara Taq Hot Start Version (Takara Biotechnology Co., Ltd.), with a total reaction volume of 25 µl (composition as above). The cycling conditions were as follows: 38 cycles of denaturation at 94°C for 30 sec, annealing at 60°C (unmethylated) or 62°C (methylated) for 30 sec and extension at 72°C for 1 min, according to the manufacturer's protocol. The post-MS-PCR products were separated on 3.0% Regular Agarose (Biowest) and analyzed using electrophoresis. DL500 DNA Marker (Takara Biotechnology Co., Ltd.) was the molecular weight marker. The following primers were used for amplification: Sense, 5′-TTAGGTTAGAGGGTTATCGCGT-3′ and antisense, 5′-TAACTAAAAATTCACCTACCGAC-3′ for a 116-bp fragment corresponding to the CDH1 methylated sequence; and sense, 5′-TAATTTTAGGTTAGAGGGTTATTGT-3′ and antisense, 5′-CACAACCAATCAACAACACA-3′ for a 97-bp fragment corresponding to the CDH1 unmethylated sequence. Regardless of whether the unmethylated allele was amplified, positivity was determined as a sample with a methylated allele. All other samples were classified as negative.
Statistical analysis
Descriptive analyses, such as mean, standard deviation, median, frequency, and percentage, were performed to explore the clinicopathological characteristics and methylation status. Pearson's χ2 test for categorical variables and Student's t-test for continuous variables were used for comparing clinical factors between tumors demonstrating hypermethylation versus no hypermethylation. Bonferroni correction was applied to adjust P-values for the multiple comparisons. Kaplan-Meier survival curves and log-rank statistics were employed to evaluate DFS and OS. Multivariate regression analysis was performed using the Cox proportional hazards model. All statistical analyses were performed using SPSS software (version 18.0; SPSS Inc., Chicago, IL, USA). Reported P-values were two-sided, and P<0.05 indicated a statistically significant difference.
Results
Patient characteristics
The present study analyzed 160 patients. At the time of diagnosis, patients with breast cancer ranged in age from 34 to 71 years old (mean age, 50.2±11.0 years; median age, 51 years). The age of patients with benign lesions ranged from 25 to 58 years (mean age, 39.8±11.9 years; median age, 38 years). Other histopathological features of the primary breast cancer patients are shown in Table I.
Table I.
Association between CDH1 methylation and clinical variables in patients with breast cancer.
| Methylation status of CDH1 | ||||
|---|---|---|---|---|
| Variable | Sample size, n | Positive, n (%) | Negative, n (%) | P-value |
| Age, years | 0.341 | |||
| ≤50 | 58 | 21 (36.2) | 37 (63.8) | |
| >50 | 79 | 35 (44.3) | 44 (55.7) | |
| Histological grade | 0.410a | |||
| I | 26 | 8 (30.7) | 18 (69.2) | |
| II | 64 | 26 (40.6) | 38 (59.4) | |
| III | 47 | 22 (46.8) | 25 (53.2) | |
| TNM stagec | 0.258b | |||
| I | 17 | 5 (29.4) | 12 (70.6) | |
| II | 81 | 32 (39.5) | 49 (60.5) | |
| III | 39 | 19 (48.7) | 20 (51.3) | |
| IV | 13 | 8 (61.5) | 5 (38.5) | |
| Lymph node metastasis | 0.022 | |||
| Positive | 77 | 38 (49.4) | 39 (50.6) | |
| Negative | 60 | 18 (30.0) | 42 (70.0) | |
| Histological type | 0.052 | |||
| Ductal | 106 | 48 (45.3) | 58 (54.7) | |
| Lobular | 31 | 8 (25.8) | 23 (74.2) | |
| ER | 0.018 | |||
| Positive | 96 | 33 (34.4) | 63 (65.6) | |
| Negative | 41 | 23 (56.1) | 18 (43.9) | |
| PR | 0.407 | |||
| Positive | 67 | 25 (37.3) | 42 (62.7) | |
| Negative | 70 | 31 (44.3) | 39 (55.7) | |
| Her-2 | 0.284 | |||
| Positive | 36 | 12 (33.3) | 24 (66.7) | |
| Negative | 101 | 44 (43.6) | 57 (56.4) | |
| p53 | 0.634 | |||
| Positive | 53 | 23 (43.4) | 30 (56.6) | |
| Negative | 84 | 33 (39.3) | 51 (60.7) | |
| Ki-67 | 0.379 | |||
| Positive | 82 | 36 (43.9) | 46 (56.1) | |
| Negative | 55 | 20 (36.4) | 35 (63.6) | |
| E-cadherin | <0.001 | |||
| Positive | 79 | 22 (27.8) | 57 (72.2) | |
| Negative | 58 | 34 (58.6) | 24 (41.4) |
CDH1, E-cadherin gene; TNM, tumor node metastasis; ER, estrogen receptor; PR, progesterone receptor; Her-2, human epidermal growth factor receptor-2; p53, tumor protein 53. Bonferroni correction was applied for statistical adjustment of each P-value due to a larger number of groups (statistically significant aP<0.017, bP<0.008). cData derived from 137 primary breast cancer patients and 13 metastatic patients. The TNM stage was the only information included in the present study for the metastatic patients, as other data for these patients were collected in various institutions and may not be accurate.
Methylation analyses
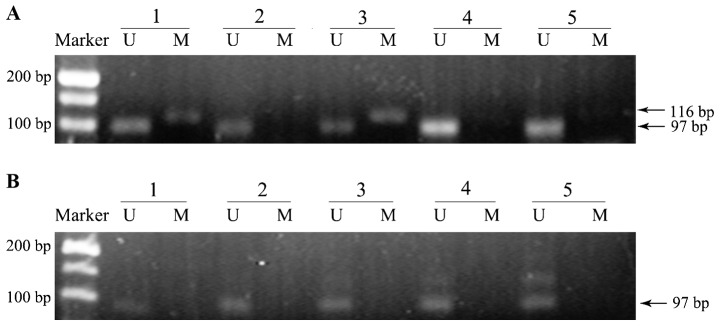
A group of representative examples of MS-PCR analysis from breast cancer and matched normal breast tissues are shown in Fig. 1. As shown, two of the five breast cancer samples exhibited promoter methylation positivity, while no methylation band was observed in matched normal tissues. The presence of a methylated band was the standard for methylation positivity.
Figure 1.
Representative example of methylation-specific polymerase chain reaction for E-cadherin gene promoter sequence in (A) breast cancer samples and (B) matched normal tissues samples. Lanes M and U correspond to specific amplification products for methylated (116 bp) and unmethylated (97 bp) alleles, respectively. Marker, DL500 DNA Marker. The presence of a methylated allele amplicon was used as standard for methylation positivity. Two of the five breast cancer samples exhibited promoter methylation and no methylation was observed in matched normal tissues.
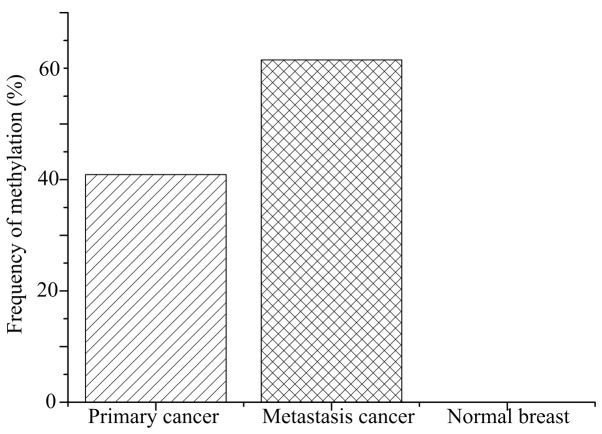
The methylation frequency of the CDH1 promoter in breast cancer tissues was 42.7% (56/137 primary cancer and 8/13 lung metastasis), significantly higher than that 0% (0/85) in the normal breast tissues (P<0.001; Fig. 2). Furthermore, the methylation status was significantly associated with lymph node metastasis (P=0.022) and ER expression (P=0.018) in patients with breast cancer. However, no statistically significant differences between CDH1 promoter methylation frequency and age (≤50 vs. >50 years), TNM stage, histological grade, histological type, PR, Her-2, p53 or Ki-67 status were identified in patients with breast cancer (Table I).
Figure 2.
Frequency of E-cadherin gene methylation in primary breast cancer (40.9%), metastatic lung cancer (61.5%) and matched normal tissues (0%). The frequency of matched normal samples was decreased compared with the other groups (P<0.001; primary cancer vs. normal breast and metastasis cancer vs. normal breast), while no significant difference was identified between the primary and metastatic cancer groups (P=0.150; primary cancer vs. metastasis cancer).
Notably, 60.7% (34/56) of primary carcinoma samples with CDH1 methylation showed negative expression of E-cadherin. However, only 29.6% (24/81) of primary carcinoma samples without CDH1 methylation showed negative expression of E-cadherin. Therefore, positive CDH1 methylation of appears to be significantly correlated with decreased E-cadherin expression (P<0.001). However, there were a number of discrepancies in the data; for example, 22 samples with positive CDH1 promoter methylation showed positive expression of E-cadherin, while 24 samples showed negative expression of E-cadherin without CDH1 methylation (Table I).
After a median follow-up of 72 months, OS and DFS data of the patients with primary breast cancer (n=137) were compared between methylated and unmethylated CDH1 promoter sequences by univariate Kaplan-Meier analysis using log-rank statistics. CDH1 methylation in primary breast cancer was significantly associated with poor OS (5-year survival: 64.3% in the methylated group vs. 80.0% in the unmethylated group, P=0.032) and DFS (5-year survival: 53.6% in the methylated group vs. 72.8% in the unmethylated group, P=0.017), as indicated by the Kaplan-Meier survival curves (Fig. 3). To verify whether CDH1 methylation was an independent prognostic factor, multivariate Cox regression analysis was conducted with various factors, including tumor size, lymph node metastasis, histological grade, and ER, PR and Her-2 status. CDH1 methylation in breast carcinoma represented an independent and strong risk factor for OS (HR, 1.737; 95% CI, 0.957–3.766; P=0.041) and DFS (HR, 2.018; 95% CI, 2.057–3.845; P=0.033) (Table II).
Figure 3.
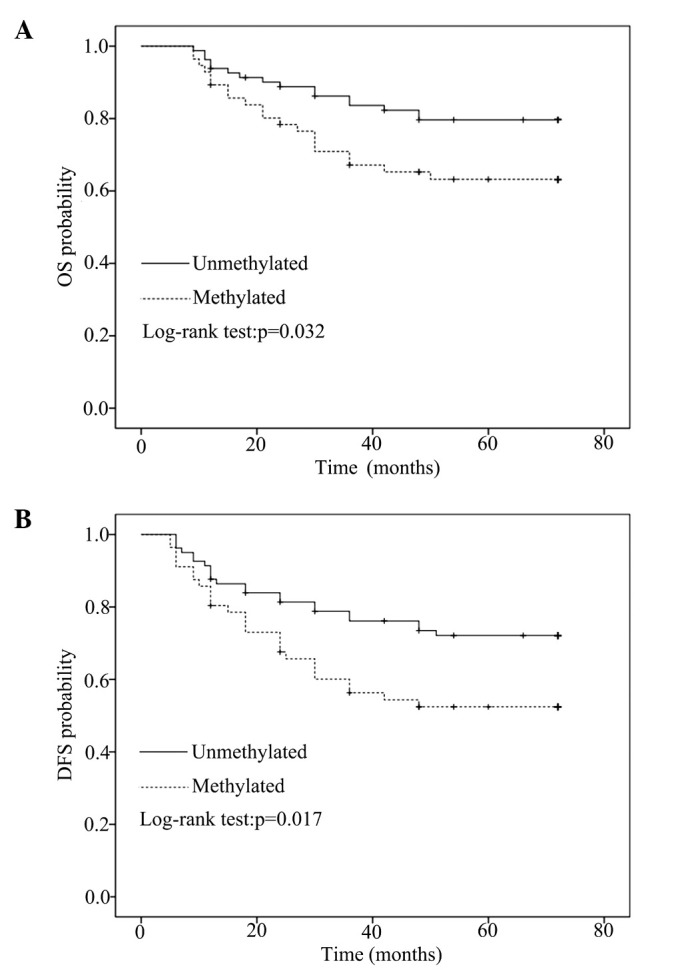
(A) OS (P=0.032) and (B) DFS (P=0.017) were analyzed by Kaplan-Meier survival analysis, according to the methylation status of the primary tumor. OS, overall survival; DFS, disease-free survival.
Table II.
Multivariate Cox regression analysis of CDH1 promoter methylation with regard to OS and DFS of patients.
| DFS | OS | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Tumor size | 0.336 | 0.074 | ||||
| T1–2 vs. T3–4 | 1.842 | 0.531–6.393 | 1.730 | 0.950–3.050 | ||
| Lymph node metastasis | 0.015 | 0.045 | ||||
| N0 vs. N1–3 | 0.294 | 0.110–0.789 | 1.937 | 0.997–3.776 | ||
| Histological grade | 0.579 | 0.305 | ||||
| I/II vs. III | 1.266 | 0.550–2.917 | 0.635 | 0.267–1.512 | ||
| ER status | 0.274 | 0.224 | ||||
| Negative vs. positive | 0.602 | 0.243–1.494 | 0.623 | 0.267–1.364 | ||
| PR status | 0.026 | 0.380 | ||||
| Negative vs. positive | 3.938 | 1.177–13.176 | 1.450 | 0.633–3.321 | ||
| Her-2 status | 0.437 | 0.763 | ||||
| Negative vs. positive | 1.580 | 1.499–5.005 | 1.134 | 0.501–2.565 | ||
| CDH1 methylation | 0.033 | 0.041 | ||||
| Negative vs. positive | 2.018 | 1.057–3.854 | 1.737 | 0.957–3.766 | ||
CDH1, E-cadherin gene; DFS, disease-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor; Her-2, human epidermal growth factor receptor-2.
Gene expression analyses
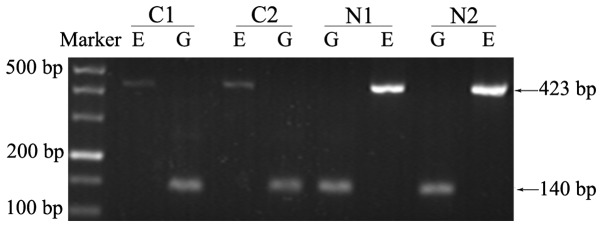
qPCR was used to quantitatively detect the relative expression level of CDH1 mRNA. The mean expression level of normal breast specimens derived from benign lesion patients was used as a baseline to calculate relative expression level change fold. By conducting RT-PCR in advance, a series of electrophoretograms were obtained to intuitively present differences in expression between cancer and normal groups (Fig. 4).
Figure 4.
Reverse transcription-polymerase chain reaction analysis of E-cadherin gene mRNA levels in breast cancer samples and matched normal tissues. The normal tissues exhibited higher expression levels of CDH1 mRNA than the matched primary cancer tissues. E, CDH1 mRNA (423 bp); G, internal control GAPDH (140 bp); C, cancer sample; N, matched normal sample.
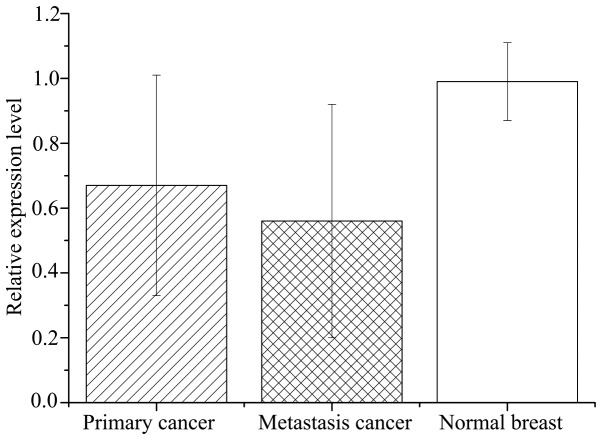
Relative expression levels of CDH1 mRNA in primary breast cancer tissues (0.67±0.34) were significantly lower than the matched normal tissues (0.99±0.12; P<0.001; Fig. 5). However, the mRNA levels varied considerably among breast cancer samples, with certain samples even exhibiting a higher expression level than the matched normal samples (Fig. 6). Relative expression of CDH1 mRNA was significantly lower in metastasis specimens (0.56±0.36) compared with primary cancer (P=0.831) and normal breast (P<0.001) tissues (Fig. 5).
Figure 5.
Relative expression level of E-cadherin gene (CDH1) mRNA was detected by reverse transcription-quantitative polymerase chain reaction analysis in primary breast cancer, lung metastatic cancer and matched normal breast tissue samples. The average expression level of CDH1 in normal breast tissue derived from patients with benign lesions was used as the control and the relative expression level was calculated as the ratio of the control. The bars and error bars represent the mean±standard deviation. The matched normal tissues exhibited higher expression (0.99±0.12) than the metastatic (0.56±0.36) and primary (0.67±0.34; P<0.001) cancer samples; however, no significant difference was observed between the primary and metastatic cancer (P>0.05). P<0.001, primary cancer vs. normal breast and metastasis cancer vs. normal breast.
Figure 6.
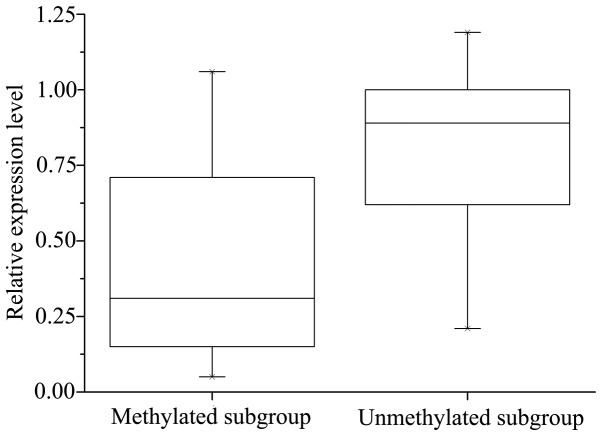
Relative expression level of E-cadherin gene messenger RNA in methylated and unmethylated breast cancer subgroup, according to reverse transcription-quantitative polymerase chain reaction. The data is presented as an interquartile range. The unmethylated subgroup exhibited higher expression level than the methylated subgroup (P=0.020). P=0.020, Methylated vs. Unmethylated subgroups.
Significant correlation was observed between CDH1 methylation and decreased expression of CDH1 mRNA: The mRNA levels of CDH1 were significantly lower in the CDH1 methylated group (0.55±0.31) compared with the unmethylated group (0.74±0.29; P=0.020). However, the methylation of CDH1 did not necessarily result in a corresponding downregulation of CDH1 mRNA expression (Fig. 6).
Discussion
The present study investigated the promoter methylation status and expression levels of CDH1 in breast cancer tissues and matched normal breast tissues. The methylation status of CDH1 was detected by MS-PCR at a sensitivity level of 1/1,000 requiring only small quantities of DNA sample (19).
In breast cancer, the incidence of CDH1 promoter methylation ranges between 21 and 72% (20–25). In agreement with this, the current study identified CDH1 promoter hypermethylation in 42.7% of patients with breast cancer (40.9% in primary cancer and 61.5% in lung metastasis). The differences in the methylation incidence observed in previous studies may result from different tissue specimen preservation (for example, fresh tissues and formalin-fixed paraffin embedded tissues). Developments in DNA methylation detection methods, such as quantitative MS-PCR, may also contribute to the variable detectable rate. Additionally, the clinical samples included in the previous studies vary in clinical stage, histological grade, histological type, metastatic status and familial breast cancer status, possibly leading to variable results.
Several previous studies have demonstrated an association between CDH1 methylation or abnormal expression of E-cadherin and breast cancer progression (23,26,27). A previous experiment in breast cancer cell lines also confirmed that CDH1 promoter methylation regulates gene expression level (14). In the present study, CDH1 methylation was correlated with the expression level of CDH1 mRNA in matched breast cancer and normal tissues. Consistent with previous studies, the incidence of CHD1 promoter methylation in the primary cancer samples (40.9%) was significantly different from that in the matched normal tissues (P<0.001), and the CDH1 mRNA levels were significantly lower than in the matched normal specimens (P<0.001). Furthermore, the current study analyzed the association between CDH1 methylation status and the expression levels in the breast cancer group. The analysis demonstrated that CDH1 methylation was significantly correlated with the downregulation of CDH1 mRNA (P=0.020) and E-cadherin expression levels (P<0.001). The aforementioned results suggest that abnormal promoter methylation is one of the mechanisms of downregulating CDH1 expression and may correlate with breast carcinogenesis.
However, CDH1 promoter methylation was not uniformly associated with the downregulation of CDH1 expression levels in the current study. In the unmethylated subgroup of the breast cancer samples, 24 samples exhibited absent expression of E-cadherin and downregulation of CDH1 mRNA. Various studies have demonstrated that CDH1 expression can be repressed by mechanisms other than promoter methylation, such as changes in chromatin structure, LOH at 16q22.1, inactivating gene mutations, specific transcriptional factors, and translational and post-translational regulation (28,29). By contrast, 5 samples of the methylated subgroup presented CDH1 mRNA expression levels that were comparable with the normal breast group, as well as positive E-cadherin expression. The presence of an unmethylated band in these specimens indicated that the high CDH1 mRNA expression level resulted from a large proportion of unmethylated cells in the specimen, and the presence of a methylated band was caused by a small number of methylated cells due to intratumoral heterogeneity.
In all samples exhibiting CDH1 methylation, unmethylated alleles invariably coexisted with methylated alleles. These unmethylated alleles may reflect the contribution of normal breast cells mixed in the samples. In addition, the unmethylated alleles may be derived from cancer cells only possessing unmethylated bases in the promoter region, as stated above. Considering that DNA methylation is a dynamic and reversible regulation mechanism, it is not unforeseen that not all cytosines in the gene promoter region were methylated or not all bases in one chain of DNA molecule were methylated (hemimethylation) by DNA methyltransmethylase 3 (DNMT3) (30). Hemimethylation also occurs during DNA replication if the maintenance mechanism of methylation based on DNMT1 is disrupted (9,31,32). Above all, the dynamic features of epigenetic regulation and complicacy of gene expression regulation are responsible for the inconformity of CDH1 promoter hypermethylation and the decreased expression of CDH1 observed in the current study.
Consistent with previous studies, the present study revealed CDH1 methylation to be significantly associated with ER-negative samples (23,33). Promoter hypermethylation is common in numerous cancer-related genes, such as RASSF1, ESR1, PGR, APC, GSTP1, BRCA, CDH13 and RARB, which also exhibit promoter hypermethylation and decreased gene expression in breast cancer (3,34,35). The correlation between ER expression and CDH1 methylation may result from the coexistence of ER and CDH1 promoter methylation, as stated in a previous study (25).
Furthermore, lymph node metastasis was identified to be significantly correlated with CDH1 methylation (P<0.05) in the current study. As patients with metastasis lesions rarely undergo surgery, only a small number of metastasis specimens were included in the present study. The incidence of methylation in metastasis was higher than that in primary cancer samples (P>0.05) and normal samples (P<0.001). The current findings indicate that CDH1 hypermethylation predominates in breast cancer cases with a more aggressive phenotype. The traditional view is that distant metastatic lesions are accompanied by a higher frequency of gene methylation, however, the majority studies do not deny the existence of unmethylated cases (36). Studies have shown that promoter hypermethylation of CDH1 independent of any gene mutation is associated with EMT and downstream events, such as aggressiveness, invasion and metastasis (37,38). Cancer cells may undergo EMT then migrate to a secondary site in the body where they occasionally undergo mesenchymal-epithelial transition (MET), reverting back to a more epithelial phenotype (39). Therefore, it should not be surprising that CDH1 hypermethylation and downregulation of CDH1 mRNA were not found in 5 distant metastasis lesions in the current study.
Notably, an association between CDH1 methylation status and clinical outcome was observed in the present study. CDH1 methylation in primary cancer indicated poor prognosis, and may be an independent prognostic factor for the OS and DFS of patients with breast cancer. However, the results may be considered weak as different postoperative treatments and a limited number of patients were included. Therefore, further investigations are required to determine the impact of CDH1 methylation on the prognosis of patients with breast cancer.
Loss of E-cadherin protein expression is most frequent in infiltrating lobular tumor types and is commonly a bi-allelic event resulting from any combination of gene promoter hypermethylation, mutation or allelic loss (40,41). While ductal cancer frequently presents with varying levels of expression, the mechanism involved in ductal breast cancer may be different from the lobular tumor types (13). However, the frequency of CDH1 methylation demonstrated no significant difference between lobular tumors and ductal tumors in the present study, although the methylation percentage in ductal cancer was larger than the lobular (P=0.052). This may be a reflection of sample selection, as the majority of cases analyzed in the present study were infiltrating ductal tumors.
No significant association was identified between CDH1 methylation, and age, clinical stage, histological grade or Her-2, PR, P53 or Ki67 status, although the majority of these parameters have been reported to be correlated with gene methylation status (21,22,25). The observation difference may result from disparate sample types. For example, patients with invasive breast cancer (stage II, grade II) accounted for majority of the total cases. Also, different standards for grouping cases based on these parameters may lead to different outcomes (21,33). Additionally, more samples and a more effective methylation detection method may aid in uncovering the association between CDH1 methylation and breast cancer.
In conclusion, the present study demonstrated that CDH1 promoter methylation may be correlated with breast carcinogenesis and indicate poor prognosis in patients with breast cancer.
Acknowledgements
The present study was supported by the Natural Science Foundation of China (approval ID: 81272418). The authors thank the assistants at the Department of Thoracic Surgery and the staff of the Key Laboratory of Environment and Genes Related to Disease (Ministry of Education, Faculty of Public Health, College of Medicine, Xi'an Jiaotong University, Xi'an, China) for their technical assistance.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2014;14:293–305. doi: 10.1586/14737159.2014.897610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol (1985) 2010;109:598–605. doi: 10.1152/japplphysiol.00066.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S. Epigenetics in breast cancer: What's new? Breast Cancer Res. 2011;13:225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitzwieser M, Holzweber E, Pfeiler G, Hacker S, Cichna-Markl M. Applicability of HIN-1, MGMT and RASSF1A promoter methylation as biomarkers for detecting field cancerization in breast cancer. Breast Cancer Res. 2015;17:125. doi: 10.1186/s13058-015-0637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews JL, Kim AC, Hens JR. The role and function of cadherins in the mammary gland. Breast Cancer Res. 2012;14:203. doi: 10.1186/bcr3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Leeuw WJ, Berx G, Vos CB, Peterse JL, Van de Vijver MJ, Litvinov S, Van Roy F, Cornelisse CJ, Cleton-Jansen AM. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J Pathol. 1997;183:404–411. doi: 10.1002/(SICI)1096-9896(199712)183:4<404::AID-PATH1148>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919–1925. [PubMed] [Google Scholar]
- 12.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Acs G, Lawton TJ, Rebbeck TR, LiVolsi VA, Zhang PJ. Differential expression of E-cadherin in lobular and ductal neoplasms of the breast and its biologic and diagnostic implications. Am J Clin Pathol. 2001;115:85–98. doi: 10.1309/FDHX-L92R-BATQ-2GE0. [DOI] [PubMed] [Google Scholar]
- 14.Benton G, Crooke E, George J. Laminin-1 induces E-cadherin expression in 3-dimensional cultured breast cancer cells by inhibiting DNA methyltransferase 1 and reversing promoter methylation status. FASEB J. 2009;23:3884–3895. doi: 10.1096/fj.08-128702. [DOI] [PubMed] [Google Scholar]
- 15.Cheng CW, Wu PE, Yu JC, Huang CS, Yue CT, Wu CW, Shen CY. Mechanisms of inactivation of E-cadherin in breast carcinoma: Modification of the two-hit hypothesis of tumor suppressor gene. Oncogene. 2001;20:3814–3823. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 16.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;41:154–161. [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertolo C, Guerrero D, Vicente F, Cordoba A, Esteller M, Ropero S, Guillen-Grima F, Martinez-Peñuela JM, Lera JM. Differences and molecular immunohistochemical parameters in the subtypes of infiltrating ductal breast cancer. Am J Clin Pathol. 2008;130:414–424. doi: 10.1309/J3QV9763DYPV338D. [DOI] [PubMed] [Google Scholar]
- 21.Sebova K, Zmetakova I, Bella V, Kajo K, Stankovicova I, Kajabova V, Krivulcik T, Lasabova Z, Tomka M, Galbavy S, Fridrichova I. RASSF1A and CDH1 hypermethylation as potential epimarkers in breast cancer. Cancer Biomark. 2011;10:13–26. doi: 10.3233/CBM-2012-0230. [DOI] [PubMed] [Google Scholar]
- 22.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 24.Tao MH, Mason JB, Marian C, McCann SE, Platek ME, Millen A, Ambrosone C, Edge SB, Krishnan SS, Trevisan M, et al. Promoter methylation of E-cadherin, p16 and RAR-β (2) genes in breast tumors and dietary intake of nutrients important in one-carbon metabolism. Nutr Cancer. 2011;63:1143–1150. doi: 10.1080/01635581.2011.605982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nass SJ, Herman JG, Gabrielson E, Iversen PW, Parl FF, Davidson NE, Graff JR. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–4348. [PubMed] [Google Scholar]
- 26.Parrella P, Poeta ML, Gallo AP, Prencipe M, Scintu M, Apicella A, Rossiello R, Liguoro G, Seripa D, Gravina C, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 27.Horne HN, Sherman ME, Garcia-Closas M, Pharoah PD, Blows FM, Yang XR, Hewitt SM, Conway CM, Lissowska J, Brinton LA, et al. Breast cancer susceptibility risk associations and heterogeneity by E-cadherin tumor tissue expression. Breast Cancer Res Treat. 2014;143:181–187. doi: 10.1007/s10549-013-2771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tryndyak VP, Beland FA, Pogribny IP. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int J Cancer. 2010;126:2575–2583. doi: 10.1002/ijc.24972. [DOI] [PubMed] [Google Scholar]
- 30.Berletch JB, Phipps SM, Walthall SL, Andrews LG, Tollefsbol TO. A method to study the expression of DNA methyltransferases in aging systems in vitro. Methods Mol Biol. 2007;371:81–87. doi: 10.1007/978-1-59745-361-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 32.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: A route to new mechanisms. EMBO Rep. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavusoglu Celebiler A, Sevinc AI, Saydam S, Canda T, Baskan Z, Kilic Y, Sakizli M. Promoter methylation and expression changes of CDH1 and P16 genes in invasive breast cancer and adjacent normal breast tissue. Neoplasma. 2010;57:465–472. doi: 10.4149/neo_2010_05_465. [DOI] [PubMed] [Google Scholar]
- 34.Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jönsson G, Olsson H, Borg A, Ringnér M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12:R36. doi: 10.1186/bcr2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathiraja TN, Stearns V, Oesterreich S. Epigenetic regulation in estrogen receptor positive breast cancer-role in treatment response. J Mammary Gland Biol Neoplasia. 2010;15:35–47. doi: 10.1007/s10911-010-9166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kominsky SL, Fackler MJ, Lahti-Domenici J, Polyak K, Sacchi N, Garrett-Mayer E, Argani P, Sukumar S. Very high frequency of hypermethylated genes in breast cancer metastasis to the bone, brain and lung. Clin Cancer Res. 2004;10:3104–3109. doi: 10.1158/1078-0432.CCR-03-0118. [DOI] [PubMed] [Google Scholar]
- 37.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, van Eijk R, Eilers PH, van de Water B, Cornelisse CJ, Cleton-Jansen AM. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial -to- mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Horssen R, Hollestelle A, Rens JA, Eggermont AM, Schutte M, Ten Hagen TL. E-cadherin promotor methylation and mutation are inversely related to motility capacity of breast cancer cells. Breast Cancer Res Treat. 2012;136:365–377. doi: 10.1007/s10549-012-2261-8. [DOI] [PubMed] [Google Scholar]
- 39.Chao YL, Shepard CR, Wells A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol Cancer. 2010;9:179. doi: 10.1186/1476-4598-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Droufakou S, Deshmane V, Roylance R, Hanby A, Tomlinson I, Hart IR. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. Int J Cancer. 2001;92:404–408. doi: 10.1002/ijc.1208. [DOI] [PubMed] [Google Scholar]
- 41.Asgeirsson KS, Jónasson JG, Tryggvadóttir L, Olafsdóttir K, Sigurgeirsdóttir JR, Ingvarsson S, Ogmundsdóttir HM. Altered expression of E-cadherin in breast cancer. patterns, mechanisms and clinical significance. Eur J Cancer. 2000;36:1098–1106. doi: 10.1016/S0959-8049(00)00062-9. [DOI] [PubMed] [Google Scholar]