Figure 1. Flow chart illustrating the experimental design of this study .
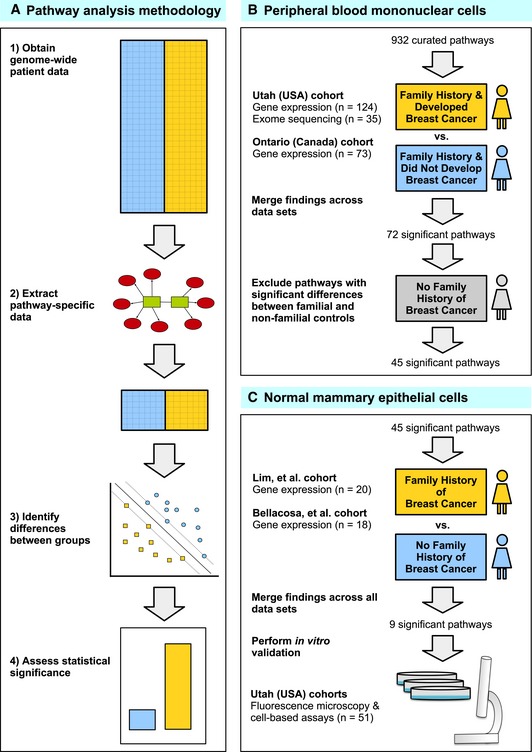
- We used pathway‐based analytic approaches to identify biological processes that may be disrupted in women who develop familial breast cancer (FBC). Having collected genomewide data, we filtered the data to include only genes associated with a given pathway. For each pathway, we identified differences between individuals who developed FBC and those who did not, using either the Support Vector Machines algorithm (gene expression data) or Barnard's exact test (DNA variant data). We considered the most statistically significant pathways to be candidates for further investigation.
- We profiled peripheral blood mononuclear cells using gene expression microarrays and exome sequencing and identified pathways that were consistently significant across these data sets. To reduce the chance that our findings were influenced by treatment effects, we excluded pathways that showed significant differences between familial and non‐familial controls.
- For the remaining pathways, we identified those that showed significant differences in two gene expression data sets representing primary mammary epithelial cells. To validate these findings, we used cell‐based assays and fluorescence microscopy to profile an additional collection of normal breast cells.
