Abstract
Individual signaling pathways are not isolated, but rather operate in the context of the broader signaling network. Thus, the response of a cell to perturbation of a given pathway depends on the state of the network, which depends upon contextual inputs from the microenvironment. The cytokine tumor necrosis factor α (TNF-α) promotes opposing cellular behaviors under different conditions, which is influenced by perturbation of the network. For example, inhibition of the mitogen-activated protein kinase (MAPK) kinase MEK alters the kinetics of TNF-α-induced apoptosis in the mouse intestinal epithelium. We investigated whether MAPK signaling directly influences TNF-α-induced apoptosis, or whether network-level effects secondary to inhibition of the MAPK pathway alter the kinetics of cell death. We found that inhibitors of the MAPK kinase kinase Raf, MEK, and extracellular signal regulated kinase (ERK) exerted distinct effects on the timing and magnitude of TNF-α-induced apoptosis in the mouse intestine. Furthermore, even different MEK inhibitors exerted distinct effects; one of them, CH5126766, potentiated TNF-α-induced apoptosis. Computational modeling analysis and experimental perturbation identified the kinase Akt as the primary signaling node that promoted apoptosis in the context of TNF-α signaling in the presence of CH5126766. Our work emphasizes the importance of integrated network signaling in specifying cellular behavior in response to external perturbation. More broadly, this study highlights the importance of considering the network-level effects of pathway inhibitors and demonstrates the distinct effects of inhibitors that share the same target.
Introduction
Cells must integrate numerous intracellular signals from a myriad of extracellular stimuli to determine their appropriate behavior. One such stimulus is tumor necrosis factor α (TNF-α), a pro-inflammatory cytokine that plays a central role in the pathogenesis of a broad range of inflammatory diseases, including inflammatory bowel diseases (IBDs) (1), rheumatoid arthritis, and psoriasis (2, 3). TNF-α impinges upon the cellular signaling network through two TNF receptors (TNFRs) that activate distinct signaling pathways: TNF-R1, which canonically stimulates a pro-death pathway through the activation of caspase-8, and TNF-R2, which canonically stimulates a pro-survival pathway through the activation of the transcription factor nuclear factor κB (NFκB) (4, 5). Because these pathways are diametrically opposed in function, TNF-α induces a broad range of cellular behaviors, including apoptosis, survival, and proliferation (6).
Previous studies have used systems analysis to rigorously identify signaling pathways that govern cell fate in cells stimulated with TNF-α. In cultured cells, mitogen-activated protein kinase (MAPK)-activated kinase 2 (MK2), NFκB, and c-Jun N-terminal kinase (JNK), are activated by TNF-α, and through a time-delayed autocrine signaling cascade to modulate the pro-apoptotic function of TNF-α (7, 8). These studies demonstrated that because the intracellular pathways downstream of the TNFRs function within the context of a broader signaling network, cell fate is determined by complex interactions within the signaling network as a whole. As such, the biological activity of TNF-α is determined by the state of the network, which is specified by factors such as genetic background, cellular differentiation state, and inputs from the extracellular environment.
The mouse intestinal epithelium provides an excellent in vivo experimental system in which to study the network-level modulation of TNF-α activity. Acute systemic exposure of a mouse to TNF-α induces apoptosis in the proximal small intestine (duodenum), but proliferation in the distal small intestine (ileum) (9). In the duodenum, the kinetics of apoptosis appear to be influenced by signaling through extracellular signal-regulated kinases 1 and 2 (ERK1/2) because the inhibition of MEK, the MAPK kinase (MAPKK) upstream of ERK1/2, accelerates the rate at which apoptosis occurs in response to TNF-α without altering the overall extent of apoptosis that occurs (9). This observation demonstrated that the biological output of TNF-α in normal cells in an intact tissue depends upon the steady state and dynamic wiring of the cellular signaling network.
Interpretation of the role of MAPK signaling in modulating TNF-α-induced apoptosis is clouded by the parallel observation that inhibition of MEK leads to broad changes to the signaling network. In addition to suppressing phosphorylation of ERK, which is the direct substrate of MEK, inhibition of MEK alters the kinetics of signaling through phosphoinositide 3-kinase (PI3K), the mammalian target of rapamycin (mTOR), and JNK in mice exposed to TNF-α (9). By extension, it is unclear whether the effect of MEK inhibition is because of the direct inhibition of ERK or is a result of secondary effects on the network. Here, we sought to address whether the MAPK pathway directly controls the kinetics of TNF-α-induced apoptosis or whether MAPK pathway inhibitors indirectly control this process. We found evidence of the latter: the effects of small molecule kinase inhibitors on the broader signaling network underlie the modulation of TNF-α-induced apoptosis in the mouse intestinal epithelium.
Results
The phosphorylation of MEK correlates with apoptosis in the TNF-α-treated mouse intestinal epithelium
We previously demonstrated that TNF-α induces apoptosis in the epithelium of the proximal small intestine over the course of four to eight hours and that inhibition of MEK with a highly-specific allosteric inhibitor alters the kinetics of this apoptotic response (9). Nevertheless, the effects of MEK inhibition on signaling were extensive, altering ERK phosphorylation as well as signaling through JNK, Akt, and S6 ribosomal protein. Based on this finding, we sought to understand whether the inhibition of MAPK signaling influenced TNF-α induced apoptosis directly or indirectly in the intestinal epithelium. We reasoned that distinct inhibitors of the MAPK pathway would similarly affect MAPK signaling, but would produce distinct effects on the rest of the signaling network because of pathway crosstalk, off-target effects, or both. If MAPK signaling directly controlled the kinetics of TNF-α-induced apoptosis, we would expect all MAPK inhibitors to exert a similar effect.
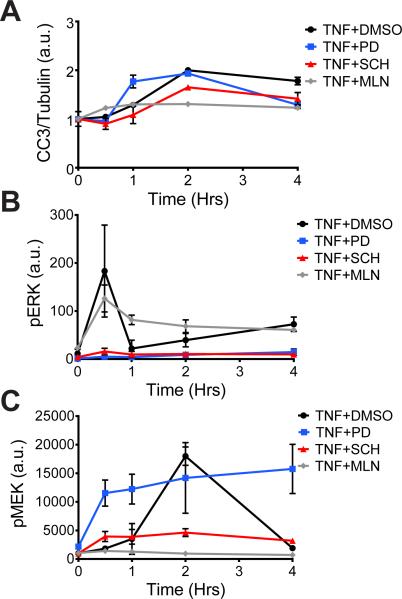
To test whether perturbation of different nodes of the MAPK pathway produced similar or distinct changes in the global signaling network and apoptotic phenotype in the intestinal epithelium, we used inhibitors of the MAPKKK Raf, MEK, and ERK. Two or four hours before mice were subjected to systemic injection with TNF-α (10), we treated them with MLN2480, a pan-Raf inhibitor, PD325901, a potent and selective MEK inhibitor, or SCH772984, an ERK1/2 inhibitor (fig. S1). At defined time points after the mice were injected with TNF-α, we quantified the amount of cleaved caspase 3 (CC3) in intestinal cell lysates as a measure of apoptosis and determined the extent of activation of various pathways by measuring the abundances of different phosphoproteins throughout the cellular network. Consistent with our previous student, we found that TNF-α induced a gradual and late (2 hours) cell death response in mice pre-treated with vehicle, whereas pre-treatment with the MEK inhibitor shifted the peak time of CC3 generation from late to early (Fig. 1A), but with <10% change in magnitude as measured by the area under the curve (AUC) (Table 1) (9). In contrast, inhibitors of Raf and ERK reduced the apoptotic response relative to that in control samples (59% and 41% of the levels of apoptosis in controls, respectively) (Fig. 1A).
Fig. 1. MAPK pathway inhibitors yield distinct phenotypic and signaling profiles in the mouse intestinal epithelium.
(A to C) Mice were pre-treated (as described in Materials and Methods) with DMSO (as a vehicle control), the MEK inhibitor PD325901 (PD), the ERK inhibitor SCH772984 (SCH), or the Raf inhibitor MLN2480 (MLN), as indicated before being treated with TNF-α for the indicated times. Intestinal epithelia from the mice were then analyzed by quantitative Western blotting or Bio-Plex to determine the relative abundance of (A) cleaved caspase-3 (CC3), normalized to that of tubulin, as an indicator of the extent of apoptosis (see Table 1); (B) pERK, normalized to that of total ERK; and (C) pMEK, normalized to that of total MEK. Data in (A) are means ± SEM of two to four experiments per time point, whereas in data in (B) and (C) are means ± SEM of three to five experiments point.
Table 1.
MAPK pathway inhibitors produce differential apoptotic responses in the mouse intestinal epithelium.
| Treatment | p-Erk | p-Mek | Relative apoptotic phenotype | CC3/tubulin area under the curve |
|---|---|---|---|---|
| DMSO | high | high | late, high | 2.52 |
| PD0325901 | low | high | early, high | 2.27 |
| SCH772984 | low | low | late, low | 1.48 |
| MLN2480 | high | low | early, low | 1.03 |
In parallel to quantifying apoptosis, we used a fluorescent bead-based enzyme-linked immunosorbent assay (ELISA) system (referred to as Bio-Plex) to determine how each of the inhibitors affected the extent of signaling through the MAPK pathway and other signaling pathways throughout the network (fig. S2). The inhibition of MEK and ERK led to the expected suppression of ERK phosphorylation; however, inhibition of Raf led to sustained ERK phosphorylation, which is a consistent finding with this drug (Fig. 1B) (11, 12). Additionally, the Raf and ERK inhibitors suppressed the phosphorylation of MEK, whereas the MEK inhibitor resulted in increased and sustained MEK phosphorylation (Fig. 1C), which is a known property of allosteric MEK inhibitors of this structural class (13, 14). Together, these results demonstrate that inhibitors targeting different nodes of the same signaling pathway can result in different phenotypic outcomes.
Mechanistically distinct MEK inhibitors produce differential apoptotic responses
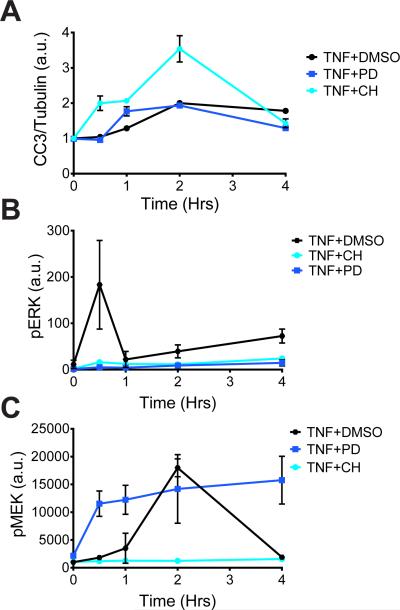
Across all of the MAPK inhibitor treatment groups, phosphorylated MEK (pMEK) was the signal that correlated best with the extent of TNF-α-induced apoptosis (Table 1). To determine whether increased MEK phosphorylation, rather than inhibition of ERK, was the key determinant of the high degree of TNF-α-induced apoptosis, we treated animals with CH5126766, an allosteric MEK inhibitor that also suppresses feedback induction of the Raf-dependent phosphorylation of MEK (14). We pre-treated mice with CH5126766 for 2 hours before they were subjected to systemic injection with TNF-α and we found that treatment with CH5126766 amplified the apoptotic response compared to that in either PD325901-treated mice or control mice (Fig. 2A). This increase in apoptosis was associated with suppressed phosphorylation of both ERK and MEK (Fig. 2 B ,C). These data suggest that the enhanced apoptotic phenotype was not a direct response to MEK phosphorylation or to the inhibition of ERK, but possibly occurred because of pathway crosstalk or off-target effects in the broader signaling network.
Fig. 2. Mechanistically distinct MEK inhibitors have differential effects on apoptosis and signaling responses.
(A to C) Mice were pretreated with DMSO, the MEK inhibitor PD325901 (PD), or the MEK inhibitor CH5126766 (CH), as indicated, before being treated with TNF-α for the indicated times. Intestinal epithelia from the mice were then analyzed by quantitative Western blotting or Bio-Plex to determine the relative abundance of (A) cleaved caspase-3 (CC3), normalized to that of tubulin; (B) pERK, normalized to that of total ERK; and (C) pMEK, normalized to that of total MEK. Data are means ± SEM of two to four experiments per time point.
Multivariate analysis identifies signaling nodes associated with apoptosis
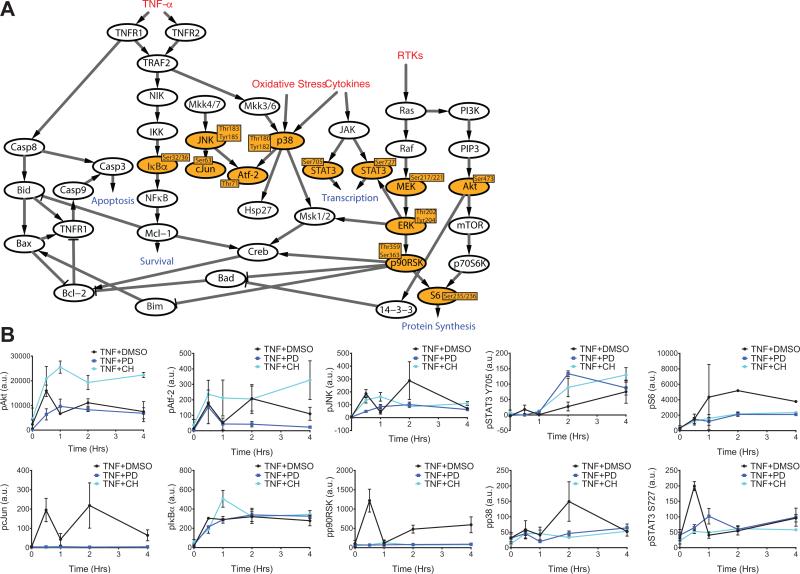
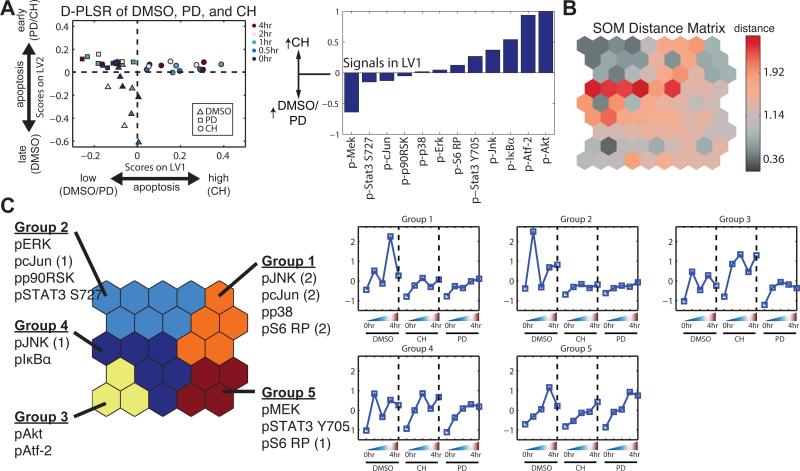
Because PD325901 and CH5126766 yielded similar changes in the abundance of pERK, but distinct apoptotic phenotypes (Fig. 2), we sought to characterize how the broader signaling network changed in response to these two MEK inhibitors. We used Bio-Plex to analyze ten phosphorylated protein (phosphoprotein) signaling nodes in addition to pERK and pMEK (Fig. 3A). PD325901 and CH5126766 exerted distinct effects on TNF-α-induced signaling through many of the pathways that we assayed (Fig. 3B). Since the relationship between signaling and biological output (that is, apoptosis) is likely to be complex, we used discriminant partial least squares regression (D-PLSR) analysis (15) to identify the signals that most strongly correlated with the enhanced apoptotic phenotype in CH5126766-treated mice. The D-PLSR analysis (Fig. 4A) incorporated all of the 12 signals that we measured. It separated the CH5126766-treated samples from the control and PD325901-treated samples along a latent variable (LV1), which was composed of a profile of signals that were increased in CH5126766-treated mice or in control and PD325901-treated mice. The signals that were most strongly associated with CH5126766 treatment (that is, enhanced apoptosis) were pAkt and pAtf-2. The signals in LV2 consisted of differences that distinguished the control mice from those treated with either of the MEK inhibitors (fig. S3A). We next used a Monte Carlo sub-sampling of 75% of the samples to construct error bars for the signals involved on the latent variables (see Materials and Methods for details). Repeating the sampling 1,000 times yielded modest standard deviations on all of the signals (fig. S3B) and coefficients of variation of 0.15 and 0.17 for pAkt and pAtf-2, respectively. Last, we re-constructed the model using all five treatment conditions to determine whether the signaling data from mice treated with the ERK inhibitor SCH772984 or the Raf inhibitor MLN2480 changed the substantial influence of pAkt and pAtf-2 on LV1. The result was a new LV1 that resembled the LV1 obtained from the original model constructed using the signaling data obtained from controls mice and those treated with either MEK inhibitor (fig. S3C and Fig. 4A).
Fig. 3. Measurement of multiple signaling nodes governing cell fate demonstrates signaling differences that are intrinsic and extrinsic to the MAPK pathway in response to PD0325901 and CH5126766.
(A) Illustration of the signaling network governing cell fate and the phosphoprotein nodes measured by Bio-Plex (highlighted in orange). (B) Mice were pretreated with DMSO, the MEK inhibitor PD325901 (PD), or the MEK inhibitor CH5126766 (CH), as indicated, before being treated with TNF-α for the indicated times. Intestinal epithelia from the mice were then analyzed by Bio-Plex to determine the relative abundances of the indicated phosphoproteins. Data in all panels are means ± SEM of two to four experiments per time point.
Fig. 4. Computational models identify signaling outside of the MAPK pathway that distinguishes the apoptotic response to MEK inhibitors.
(A) Left: D-PLSR analysis bygrouping all time points and replicates from each condition revealed an axis (LV1) thatdistinguishes the CH5126766-treated condition from the DMSO- and PD325901-treatedconditions. Right: LV1 is composed of signals (phosphoproteins) that were increased or decreased in abundance in the CH5126766-treated conditions relative to those in the DMSO- and PD325901-treated conditions. (B) An SOM was used to identify signaling pathways that clustered across temporal and conditional space. The SOM produced a nodal distance matrix that yielded at least three clear valleys: upper left, upper right, and lower left. (C) A k-means clustering of the distance matrix. Each signaling group corresponds to a distinct temporal activation profile for each condition. Group 3, which corresponds to Akt and Atf-2 signaling, is highly up-regulated in CH relative to both the DMSO and PD conditions. Signals projected into different groups from replicates (1) and (2) (fig. S3B) are indicated.
Although the D-PLSR analysis identified signals that distinguished CH5126766-treated mice from control mice and those treated with PD325901, it did not take into account the kinetic nature of the changes in the phospho-protein signaling network. To address this limitation, and to confirm the strong association between enhanced apoptosis and the phosphorylation of Akt and Atf-2, we used a self-organizing map (SOM) to gain clearer insight into which signals changed together as a group, and how each group changed over time and across administered conditions (16, 17). An SOM consists of a two-dimensional matrix of nodes, each of which contains a vector of coefficients that is the same length as the data vectors being presented to the map for clustering. The nodal coefficient vectors are tuned to optimally capture the domain of the vectors being presented, with similar nodes appearing near each other in the tuned SOM (16). An SOM consists of a two-dimensional matrix of nodes, each of which contains a vector of parameters that is the same length as the data vectors that are presented to the map for clustering. The parameters in the various nodes are tuned to optimally mirror the collection of data point vectors presented, with similar nodes being adjacent to one another and very different nodes being on opposite sides of the map (16). By assembling a separate data vector for each signal, which is composed of the signaling value for each condition and time point, we can analyze which signals are co-varying across condition and time.
To build the map, we first concatenated all 12 signal measurements across time and condition. Each data point presented to the SOM was an aggregate vector consisting of a phosphoprotein signal collected from each of 15 mice (3 conditions and 5 time points). We measured 12 signals in total from the same 15 mice, thus generating 12 individual data points to present to the model. We obtained these vectors in biological duplicate, so we presented a total of 24 phosphoprotein data points to the SOM for training (fig. S4A). The SOM consisted of a grid of 5×5 nodes. Each node consisted of a vector of 15 parameters, with one parameter for each condition and time point in the 24 presented data point vectors. The product was a unified distance matrix that enabled us to visualize the similarity between adjacent nodes (Fig. 4B). Little distance between nodes (gray) means the nodes are similar, so a data point projecting onto either node is roughly equivalent and these nodes can be grouped. Adjacent nodes separated by greater distance (red) are relatively dissimilar and should not be grouped. Each gray region, which is not separated by red, defines a cluster of nodes that coordinately varied across time and condition. The trained SOM identified a temporal and conditional progression of nodal parameters for each signal (fig. S4B).
A k-means clustering of the nodal distance matrix identified five distinct groupings in the SOM, which mapped to groups of phosphoproteins (Fig. 4C, left). Averaging of the nodes associated with each group yielded temporal and conditional activation profiles for each group (Fig. 4C). Groups 1, 2, and 5 define signaling differences between the DMSO-treated mice and the mice treated with either CH5126766 or PD325901, such as in the phosphorylation of ERK and JNK, whereas Group 4 defines signaling that was similar across all conditions. Note that replicates of some of the signals (p-c-Jun, p-JNK, and p-S6 RP) projected onto different groups: Group 1 for replicate (2) vs. Groups 2, 4, and 5 for replicate (1). Groups 1, 2, and 5 are similar and all reflect the signaling observed in the DMSO-treated mice. The differences in group and projection position were a result of differences in the magnitude and phase of these signals between replicates in the DMSO-treated group (fig. S4A). Furthermore, the SOM showed that both replicates of pAkt and pAtf-2 projected into the same Group and that CH5126766 resulted in coordinated increases in the abundances of pAkt and pAtf-2 (Group 3), compared with DMSO and PD325901. Together with our re-sampling analysis of the D-PLSR model, the SOM supports the notion that these signals were co-activated in the CH5126766-treated mice. Finally, as with the D-PLSR model, an SOM model produced with all five available conditions (DMSO, PD, CH, MLN, and SCH) resulted in clustering and a CH5126766-dependent activation profile implicating pAkt and pAtf-2 in enhanced apoptosis, similarly to the original three-condition SOM (fig. S4C and Fig. 4C),. These models together emphasize the connection between Akt and Atf-2 signaling and the highly apoptotic phenotype observed in the CH5126766-treated mice.
Co-application of CH5126766 and Akt inhibitors reduces TNF-α-induced apoptosis
The D-PLSR and SOM analysis identified pAkt and pAtf-2 as the factors that correlated best with the enhanced apoptotic phenotype seen in the CH5126766-treated animals. Because Atf-2 inhibitors are not available, we tested whether inhibition of the PI3K pathway could alter the apoptotic phenotype. To perturb signaling upstream and downstream of Akt, we first treated mice with a combination of the dual PI3K and mTOR inhibitor BEZ235 (18) and either PD325901 or CH5126766. The combination treatment of BEZ235 and PD325901 led to a slight increase in apoptosis relative to that observed in mice treated with PD325901 alone; however, the combination of BEZ235 and CH5126766 substantially decreased the extent of apoptosis relative to that observed in mice treated with CH5126766 alone (Fig. 5A).
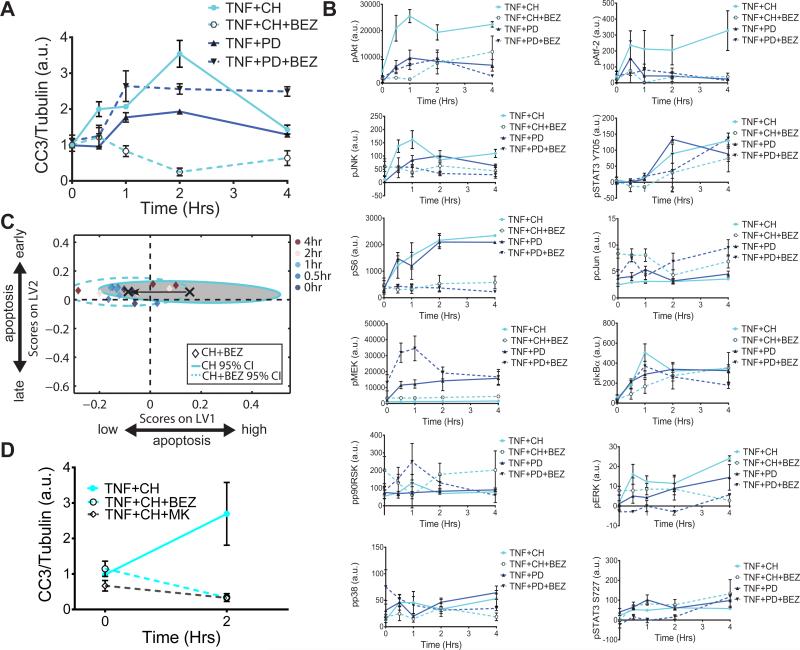
Fig. 5. Global network shifts govern the apoptotic response to the combinatorial application of MEK and Akt inhibitors.
(A and B) Mice were pretreated with the MEK inhibitor PD325901 (PD) or the MEK inhibitor CH5126766 (CH) in the presence or absence of the PI3K inhibitor NVP-BEZ235 (BEZ), as indicated, before being treated with TNF-α for the indicated times. Intestinal epithelia from the mice were then analyzed by quantitative Western blotting or Bio-Plex to determine (A) the relative abundance of cleaved caspase-3 (CC3), normalized to that of tubulin, as an indicator of apoptosis, and (B) the relative abundances of the indicated phosphoproteins. Data in (A) are means ± SEM of three experiments per time point, whereas data in (B) are means ± SEM of three or four experiments per time point. (C) The computational D-PLSR model derived from the DMSO, CH5126766-treated, and PD325901-treated conditions (from Fig. 3B). X's and ellipses represent centroids and 95% confidence intervals (CIs) (34) for the CH and CH+BEZ treatment groups, respectively. (D) Mice were pretreated with the MEK inhibitor CH5126766 (CH) in the presence or absence of the PI3K inhibitor NVP-BEZ235 (BEZ) or the Akt inhibitor MK2206 (MK), as indicated, before being treated with TNF-α for the indicated times. Intestinal epithelia from the mice were then analyzed by quantitative Western blotting or Bio-Plex to determine the relative abundance of cleaved caspase-3 (CC3), normalized to that of tubulin. Data are means ± SEM of two to five experiments per time point.
Whereas, BEZ235 reduced the abundances of both pAkt and pAtf-2 compared to those in mice treated with DMSO (fig. S5), the combined treatment of mice with BEZ235 and PD325901 only slightly reduced the amounts of pAkt and pAtf-2 relative to those DMSO-treated controls (Fig. 5B), which suggests that the response of the signaling network to pharmacological intervention was context dependent. Furthermore, CH5126766 alone resulted in decreased amounts of both pAkt and pAtf-2 (Fig. 5B), which is suggestive of a non-canonical relationship between Akt phosphorylation and apoptosis in CH5126766-treated cells.
Because, as with all of the MAPK pathway inhibitors, PI3K inhibition was associated with other broad network changes, we next used our original D-PLSR model (Fig. 4A) to determine whether the broad signaling changes observed with co-administered CH5126766 and BEZ235 would predict reduced apoptosis. Projection of the signaling data obtained from mice treated with CH5126766 and BEZ235 onto the original computational model demonstrated a clear shift from the high- to low-apoptosis region of the scores plot (Fig. 5C).
Finally, to verify that the apoptotic phenotype was Akt-dependent, we treated mice with a combination of the highly specific Akt inhibitor (MK-2206) (19) and CH5126766 (Fig. 5D). This co-treatment resulted in a decrease in the extent of apoptosis that was similar to that elicited by the combination of CH5126766 and BEZ235, as well as a similar reduction in the abundances of pAkt and pAtf-2 (fig. S4C). These results raise the possibility that the normally pro-survival Akt pathway may instead promote apoptosis in the context of TNF-α signaling in the presence of CH5126766, a MEK inhibitor that suppresses feedback activation of Raf.
Discussion
We previously used the mouse intestine as a model system in which to study how cell-autonomous and non-cell-autonomous signaling pathways modulate the biological activity of TNF-α in an intact tissue (9, 20). A key finding from these previous studies was that inhibition of MEK alters the kinetics of TNF-α-induced apoptosis in the intestinal epithelium without altering the overall extent of apoptosis (9). The straight forward interpretation of this observation – that loss of ERK activation downstream of MEK accounts for the change in apoptotic phenotype – is confounded by the fact that MEK inhibition affects the phosphorylation of many proteins after TNF-α stimulation. We hypothesized, therefore, that the network-level effects of MEK inhibition might alter the kinetics of TNF-α-induced apoptosis. By extension, different drugs acting on the MAPK pathway could differentially affect cell fate based on the direct and indirect activities of each drug. We tested our hypothesis by administering to mice drugs that inhibit different nodes of the presumably linear MAPK pathway. These inhibitors yielded a range of apoptotic phenotypes in animals treated with TNF-α, despite the ability of all inhibitors to alter the kinetics of ERK phosphorylation (Fig. 1). This observation provided the first clue that flux through the MAPK pathway is not directly responsible for modulating the pro-apoptotic activity of TNF-α in the intestinal epithelium.
Although the kinetics of ERK phosphorylation did not correlate with the apoptotic phenotypes that resulted from inhibition of the MAPK pathway, we found that MEK phosphorylation, which was increased in TNF-α-treated control animals and in those treated with the MEK inhibitor PD325901, correlated with a relatively high incidence of apoptosis (Table 1). We attempted to validate the hypothesis that pMEK was pro-apoptotic in experiments with a second MEK inhibitor (CH5126766), which inhibits the pathway without enhancing MEK phosphorylation, but this treatment strongly potentiated TNF-α-induced apoptosis (Fig. 2). To understand that broad impact of different MEK inhibitors on the cellular signaling network, we used multiple computational modeling approaches to identify signals outside of the MAPK pathway that correlated with the different apoptotic phenotypes induced by PD0325901 and CH5126766 (Fig. 4). Each of the computational approaches pointed toward a pro-apoptotic signaling axis that includes Akt and Atf-2.
The limitation of the computational modeling approaches that we used is that they are merely correlative. As such, formal validation of the role of Akt and Atf-2 in TNF-α-induced apoptosis required experimental validation. To this end, we co-administered CH5126766 to mice together with a PI3K and mTOR inhibitor (BEZ235) or an allosteric Akt inhibitor (MK-2206) to test whether inhibition of Akt would reverse or enhance the pro-death phenotype caused by CH5126766. Treatment with both BEZ235 and MK-2206 resulted in a substantial decrease in the extent of apoptosis, relative to that observed in mice treated with CH5126766 alone (Figs. 5A), suggesting that Akt is a causal node driving apoptosis. Re-analysis of the multi-pathway signaling network response after co-delivery of CH5126766 and BEZ235 showed that the abundances of pAkt and pAtf-2 were reduced to amounts similar to those in mice treated with either vehicle or PD325901, which did not induce apoptosis to a great extent (Fig. 5B). To our knowledge, a direct link between Akt and Atf-2 signaling has not been reported, but our data suggest a possible relationship between these two pathways, potentially mediated through other measured (for example, c-Jun) or unmeasured signaling nodes. Furthermore, co-treatment with PD325901and BEZ235 resulted in increased apoptosis relative to that in mice treated with PD325901 alone (Fig. 5A), which was associated with network changes not observed in the CH5126766 and BEZ235 co-treatment (Fig. 5B). Although co-treatment of mice with PD325901and MK-2206 would further validate a context-dependent pro-apoptotic role of Akt, we found this treatment combination to be highly toxic, leading to premature death in mice. This finding demonstrates that different inhibitors of the same enzyme can exhibit distinct interactions with other inhibitors, depending on the context in which they are combined. This finding is particularly important given the current clinical evaluation of combined MAPK and PI3K therapies for cancer treatment (21-23).
Akt is a canonically pro-survival signal (24, 25), and our finding that Akt was causative for, rather than responsive to, apoptosis was surprising; however, a pro-death role for Akt has been reported in several contexts. Strong Akt activation induces cellular senescence and increases oxidative stress, rendering cells susceptible to reactive oxygen species (ROS)-induced cell death (26). Cells over-expressing the oncoprotein Bcr-Abl, which causes enhanced activation of Akt, have increased sensitivity to ROS (27). Akt activates apoptosis-regulating kinase 1 (Ask1) to promote apoptosis in response to ROS (28). Moreover, while the canonical role of Akt is at the plasma membrane, all components of the PI3K pathway are also found in the nucleus; thus, nuclear Akt may phosphorylate substrates that are distinct from those that it targets in the cytoplasm. Consistent with this idea, the enhanced activation of nuclear Akt by Apoptin, a viral protein that is considered to have anti-cancer properties, potentiates cell death (29, 30). Our findings therefore contribute to the growing body of literature that indicates a context-specific pro-death role for Akt signaling.
Why does CH5126766 strongly activate Akt, whereas PD325901 does not? Indeed, PD325901 suppressed the initial burst of Akt phosphorylation that is induced by TNF-α (Fig. 3A). CH5126766 and PD325901 are both allosteric inhibitors with high specificity for MEK, so the difference in their ability to promote TNF-α-induced apoptosis is likely to be “on-target,” at least with respect to its initial mechanism of action. Nevertheless, if the pro-apoptotic effect of CH5126766 is related to binding to MEK, it cannot be linked to the kinase activity of MEK, because both CH5126766 and PD325901 had the same effect on ERK phosphorylation (Fig. 2B). The primary difference between CH5126766 and PD325901 is that CH5126766 also inhibits the kinase activity of Raf by preventing the phosphorylation and release of MEK from the Raf-MEK complex (14). This indirect inhibitory activity could play a role in the specific enhancement of TNF-α-induced apoptosis by CH5126766; however, this is unlikely to be related to the kinase activity of Raf given that the direct inhibition of Raf with MLN2480 reduced, rather than enhanced, the phosphorylation of Akt and the extent of apoptosis (Fig. 1A, fig. S2). Alternatively, because CH5126766 locks Raf and MEK into an inactive complex, the drug may affect a function of Raf (or MEK) that is not related to kinase function. Indeed, many kinases have scaffolding functions that are independent of their kinase activities (31). Although kinase-independent functions of MEK have not been reported, Raf-1 functions as an inhibitor of Ask1, a role that does not require Raf kinase activity (32). Based on our data, we favor a model in which Raf or MEK inhibits the TNF-α-induced activation of Akt in a kinase-independent manner, and that this function is inhibited by CH5126766, but not by PD325901. CH5126766 therefore activates Akt and promotes TNF-α-induced apoptosis in terminally differentiated intestinal epithelial cells in vivo.
Pharmacologic interventions in human diseases are delivered to a complex tissue microenvironment, which plays a major role in establishing the disease-specific signaling network. TNF-α is a prime example of a factor that is found in the local and systemic environments of many diseases, including cancer (1, 4). The multiple pathways that TNF-α stimulates shift the global signaling network (7, 8), which in turn determines cellular behaviors within a tissue. Furthermore, the global signaling network determines not only the phenotypic outcome in response to TNF-α, but also determines how the tissue will respond to additional perturbations, such as pharmacologic interventions. Whereas other groups have evaluated network-level signaling effects of TNF-α (7, 8, 33), we systematically evaluated how both phenotype and global signaling changed in response to multiple perturbations to the MAPK pathway in the context of TNF-α signaling, and how both phenotype and signaling change in response to the co-inhibition of Akt. The possibility that Akt acts as a pro-death signal in the context of TNF-α signaling suggests that inhibitors targeting both the MAPK and PI3K pathways may reduce the extent of apoptosis more substantially than would a therapy targeting the MAPK pathway alone, especially in tumors that have substantial inflammation. By extension, CH5126766 may be particularly effective, relative to other Mek inhibitors, as a monotherapy in cancers associated with increased amounts of TNF-α. More generally, our work demonstrates that the network level signaling must to be considered in the development of new combinatorial strategies.
Materials and Methods
Mice, inhibitor treatments, and tissue isolation
All mouse work performed in this study was completed according to a protocol approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. All mice were 8-week old males on the C57BL/6J genetic background (Jackson Laboratory). For the MEK inhibition experiments, mice were anesthetized with Avertin (tribromoethanol, 250 mg/kg), and then subjected to retro-orbital injection with PD325901 (12.5 mg/kg, ChemieTek) solubilized with 10% dimethyl sulfoxide (DMSO) in PBS or to intraperitoneal (IP) injection with CH5126766 (2.0 mg/kg, Active Biochemicals) solubilized with 10% DMSO in PBS. For the ERK inhibition experiments, mice were subjected to IP injection with SCH772984 (75mg/kg, Selleckchem) solubilized with cyclodextrin in PBS. For the Raf inhibition experiments, mice were subjected to oral gavage with MLN2480 (12.5 mg/kg, Millennium) solubilized with 100% PEG400. For PI3K inhibition experiments, animals were subjected oral gavage with BEZ235 (45 mg/kg, LC Laboratories) solubilized in 10% DMSO in PBS. For Akt inhibition experiments, animals were subjected to oral gavage with MK-2206 (240 mg/kg, ChemieTek) solubilized in 15% DMSO in water. Two or four hours later, mice were anesthetized and subjected to retro-orbital injection with 5 μg of recombinant mouse TNF-α (Abazyme) solubilized in PBS. The mice were then sacrificed according to the defined time course after treatment with TNF-α. Control animals were pretreated with vehicle two or hour hours before they were injected with TNF-α. For tissue collection, mice were sacrificed and the small intestine was removed and washed with cold PBS. Tissue was lysed and homogenized in Bio-Plex lysis buffer (Bio-Rad) and stored at −80°C. The proximal small intestine (duodenum) was obtained from tissue immediately adjacent to the stomach.
Western blotting
Polyacrylamide gels (12%) were loaded with 45 μg of mouse epithelial intestinal tissue. Western blotting analysis was performed with rabbit antibody against cleaved caspase 3 (CC3, Cell Signaling Technology) and mouse antibody against α-tubulin (Sigma-Aldrich). Membranes were incubated with primary antibodies overnight at 4°C and then were incubated for 1 hour at room temperature with secondary antibodies (Rockland) at a 1:10,000 dilution. Membranes were scanned with a LI-COR Odyssey infrared imaging system. Bio-Plex signaling analysis was performed on the same tissue lysates that were used for the Western blotting analysis of CC3. The area under the curve (AUC) was computed in Prism 6 software (Graphpad Software, Inc.) with a trapezoid rule and a baseline of y = 1.
Bio-Plex signaling quantification
Quantitative measurements of protein phosphorylation in lysed issue samples were obtained with Bio-Plex phospho-signaling assay kits (Bio-Rad) for: pAkt (Ser473), pAtf-2 (Thr71), pc-Jun (Ser63), pERK1 (Thr202 and Tyr204) and pERK2 (Thr185 and Tyr187), pMEK1 (Ser217 and Ser221), pp38 (Thr180 and Tyr182), pp90Rsk (Thr359 and Ser363), pS6 (Ser235 and Ser236), phosphorylated inhibitor of nuclear factor κB α (pIκβα, Ser32 and Ser36), pJNK (Thr183 and Tyr185), pSTAT3 (Ser727), and pSTAT3 (Tyr705). Tissue lysates were quantified by BCA (bicinchoninic acid, Pierce) and equal amounts of total protein from each sample were used for each Bio-Plex signal across all samples. In particular, 4 μg of protein was used for assays of pAkt, pAtf-2, pJNK, and p-STAT3 (Tyr705) assays, and 1.5 μg of protein was used for each of the rest of the assays. These total protein amounts were previously determined to be within the linear range of each signal in small intestinal tissue samples (9). To calibrate signal measurements between Bio-Plex runs, five samples were replicated, and used to generate a linear calibrate curve between runs for each signal.
D-PLSR analysis
All D-PLSR model analysis was conducted in MATLAB (Mathworks) with the partial least squares algorithm by Cleiton Nunes (available on the Mathworks File Exchange). All Bio-Plex signaling data were z-scored, and then used as the independent (X) inputs to the algorithm. An orthogonal rotation in the LV1-LV2 plane was used to choose a new LV1 that best separated the CH5126766-treated samples from the DMSO- and PD325901-treated samples. 95% confidence ellipsoids were computed with the inverse χ2 distribution with two degrees of freedom (34). A Monte Carlo sub-sampling with 1,000 iterations was used to characterize the standard deviation (SD) of the individual signals involved in LV1 and LV2 of the D-PLSR model constructed from the DMSO, CH5126766-treated, and PD325901-treated conditions (the total model). For each iteration, 75% (28/38) of the samples used to construct the total D-PLSR model were randomly sampled, and a new D-PLSR model was constructed. To correct for sign reversals, each of the two sub-sampled LV1 and LV2 was multiplied by the sign of the scalar product of the new LV and the corresponding LV from the total model. The same orthogonal rotation used for the total model was applied to the LVs from each iteration, and the mean and SD was computed for each signal across all iterations.
SOMs
The SOM was generated with the SOM Toolbox 2.0 (35) in MATLAB. The data for each signal were concatenated across condition and time. Because a complete set of conditions and time measurements was needed for each signal sample, the number of concatenated replicates for each signal was the minimum number of samples taken for each condition and time point. Because the minimum n = 2 across all conditions and time points, N = 2 for the concatenated samples for each signal. The concatenations were assembled from the first two samples from each condition and time point. The data for each signal were z-scored across condition and time, and directly inputted to the SOM algorithm. As recommended (35), the number of nodes, m, was heuristically selected to be approximately equal to , where l is the number of data points presented to the map for training. The SOM was trained through the batch method with a Gaussian neighborhood function. A k-means clustering was used to assign each node to a group. As recommended (36), the maximum number of clusters, k, that was considered was taken to be . The Davies-Bouldin index (37) revealed that the best clustering was achieved when k=5.
Supplementary Material
Acknowledgments
We thank A. Toker for helpful comments on the manuscript. Funding: This work was supported by National Institutes of Health grant R01-GM088827 to K.M.H. and D.A.L. and American Cancer Society postdoctoral fellowship PF-13-081-01-TBG to J.J.G..
Footnotes
Author contributions: K.M.H., and D.A.L. designed the study and supervised the work. J.J.G, C.G., K.L, and Y.L. performed the mouse experiments. J.J.G and L.B.W performed the Bio-Plex assays. A.A.S. provided reagents. L.B.W conducted the computational analysis. J.J.G, L.B.W, and K.M.H wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. New Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clancy RM, Marion MC, Kaufman KM, Ramos PS, Adler A, G. International Consortium on Systemic Lupus Erythematosus. Harley JB, Langefeld CD, Buyon JP. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus. Arthritis Rheum. 2010;62:3415–3424. doi: 10.1002/art.27658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119:651–665. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters JP, Pober JS, Bradley JR. Tumour necrosis factor and cancer. J Pathol. 2013;230:241–248. doi: 10.1002/path.4188. [DOI] [PubMed] [Google Scholar]
- 6.Schrofelbauer B, Hoffmann A. How do pleiotropic kinase hubs mediate specific signaling by TNFR superfamily members? Immunol Rev. 2011;244:29–43. doi: 10.1111/j.1600-065X.2011.01060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 8.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 9.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 12.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. Journal Biological Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 14.Ishii N, Harada N, Joseph EW, Ohara K, Miura T, Sakamoto H, Matsuda Y, Tomii Y, Tachibana-Kondo Y, Iikura H, Aoki T, Shimma N, Arisawa M, Sowa Y, Poulikakos PI, Rosen N, Aoki Y, Sakai T. Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Res. 2013;73:4050–4060. doi: 10.1158/0008-5472.CAN-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi-and megavariate data analysis. Umetrics; 2006. [Google Scholar]
- 16.Kohonen T. The self-organizing map. Neurocomputing. 1998;21:1–6. [Google Scholar]
- 17.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim HD, Grantcharova V, Lauffenburger DA, White FM. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Systems Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria C. Identification and characterization of NVPBEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 19.Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan BS, Kotani H. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Tther. 2010;9:1956–1967. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 20.Lau KS, Cortez-Retamozo V, Philips SR, Pittet MJ, Lauffenburger DA, Haigis KM. Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-alpha-induced apoptosis in the intestinal epithelium. PLoS biology. 2012;10:e1001393. doi: 10.1371/journal.pbio.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franz DN, Weiss BD. Molecular therapies for tuberous sclerosis and neurofibromatosis. Curr Neurol Neurosci Rep. 2012;12:294–301. doi: 10.1007/s11910-012-0269-4. [DOI] [PubMed] [Google Scholar]
- 22.Grant S. Cotargeting survival signaling pathways in cancer. J Clin Invest. 2008;118:3003–3006. doi: 10.1172/JCI36898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, Basecke J, Stivala F, Donia M, Fagone P, Malaponte G, Mazzarino MC, Nicoletti F, Libra M, Maksimovic-Ivanic D, Mijatovic S, Montalto G, Cervello M, Laidler P, Milella M, Tafuri A, Bonati A, Evangelisti C, Cocco L, Martelli AM, McCubrey JA. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–164. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandel ES, Skeen J, Majewski N, Di Cristofano A, Pandolfi PP, Feliciano CS, Gartel A, Hay N. Activation of Akt/protein kinase B overcomes a G(2)/m cell cycle checkpoint induced by DNA damage. Mol Cell Biol. 2002;22:7831–7841. doi: 10.1128/MCB.22.22.7831-7841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 26.Nogueira TC, Anhe GF, Carvalho CR, Curi R, Bordin S, Carpinelli AR. Involvement of phosphatidylinositol-3 kinase/AKT/PKCzeta/lambda pathway in the effect of palmitate on glucose-induced insulin secretion. Pancreas. 2008;37:309–315. doi: 10.1097/mpa.0b013e318168dac3. [DOI] [PubMed] [Google Scholar]
- 27.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Pan J, Chang Q, Wang X, Son Y, Zhang Z, Chen G, Luo J, Bi Y, Chen F, Shi X. Reactive oxygen species-activated Akt/ASK1/p38 signaling pathway in nickel compound-induced apoptosis in BEAS 2B cells. Chem Res Toxicol. 2010;23:568–577. doi: 10.1021/tx9003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddika S, Wiechec E, Ande SR, Poon IK, Fischer U, Wesselborg S, Jans DA, Schulze-Osthoff K, Los M. Interaction with PI3-kinase contributes to the cytotoxic activity of apoptin. Oncogene. 2008;27:3060–3065. doi: 10.1038/sj.onc.1210958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddika S, Panigrahi S, Wiechec E, Wesselborg S, Fischer U, Schulze-Osthoff K, Los M. Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin's anticancer toxicity. Mol Cell Biol. 2009;29:1235–1248. doi: 10.1128/MCB.00668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch J, Volinsky N, Romano D, Kolch W. The secret life of kinases: functions beyond catalysis. Cell Commun Sig. 2011;9:23. doi: 10.1186/1478-811X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci USA. 2001;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wynn ML, Ventura AC, Sepulchre JA, Garcia HJ, Merajver SD. Kinase inhibitors can produce off-target effects and activate linked pathways by retroactivity. BMC Syst Biol. 2011;5:156. doi: 10.1186/1752-0509-5-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worley B, Halouska S, Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem. 2013;433:102–104. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vesanto J, Himberg J, Alhoniemi E, Parhankangas J. SOM toolbox for Matlab 5. Citeseer; 2000. [Google Scholar]
- 36.Vendramin L, Campello RJ, Hruschka ER. SDM. SIAM; 2009. pp. 733–744. [Google Scholar]
- 37.Davies DL, Bouldin DW. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell. 1979;1:224–227. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.