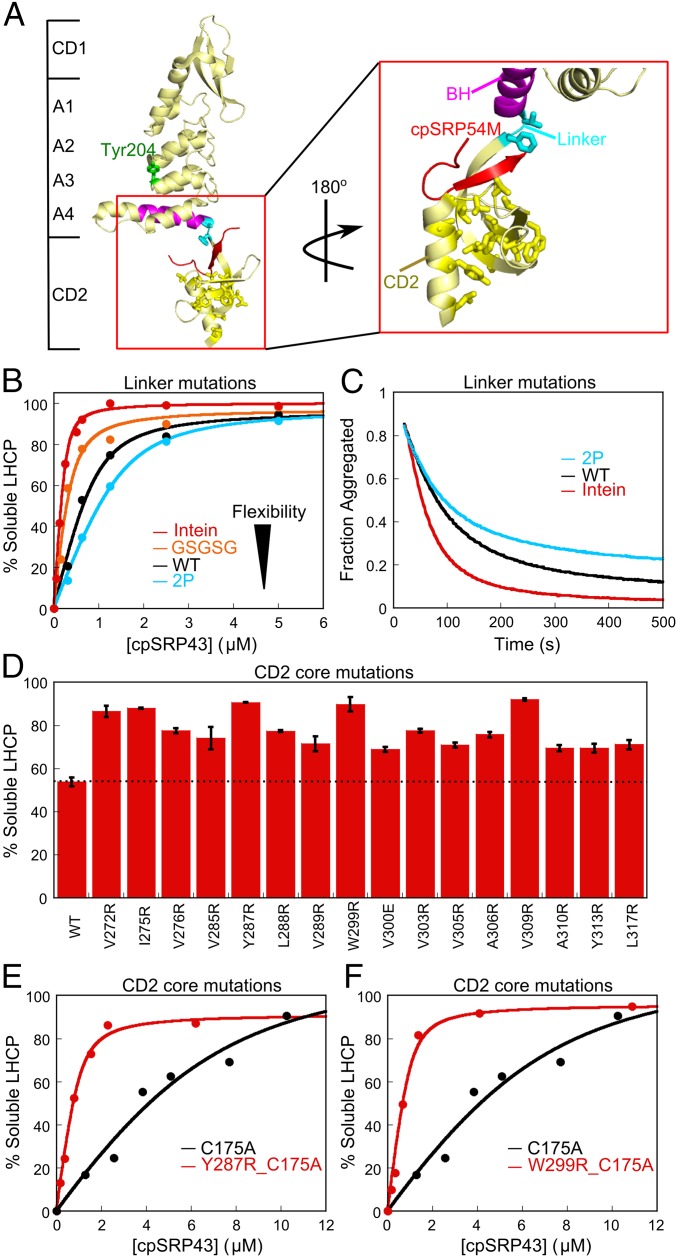
Fig. 2.
Mutations in CD2 hyperactivate the chaperone. (A) Crystal structure of cpSRP43 CD1Ank-BH-CD2 fragment bound to 54M peptide [Protein Data Bank (PDB) ID code 3UI2]. Cyan shows the linker (V266, F267) between BH and CD2, yellow shows residues mutated in the hydrophobic core of CD2, magenta highlights the BH, and red shows the 54M peptide. (B and C) Chaperone activity of cpSRP43 linker mutants, measured by prevention (B) and reversal (C) of LHCP aggregation. The data in B were fitted to Eq. 1 and gave apparent Kd values of 33, 100, 177, and 256 nM for LHCP binding to intein-cpSRP43 (red), GSGSG-cpSRP43 (orange), WT cpSRP43 (black), and 2P-cpSRP43 (cyan), respectively. (D) Many point mutations in the conserved hydrophobic core of CD2 activate chaperone activity. Chaperone activities were measured by prevention of LHCP aggregation using 1 µM LHCP and 4 µM WT or mutant cpSRP43. Error bars represent SD, with n = 2. To rule out potential involvements of CD3, all measurements were made with the ΔCD3 construct. A soft mutation, C175A, was present in all constructs to increase the sensitivity of the mutational screen under these conditions. (E and F) Mutants Y287R (E) and W299R (F) enhance LHCP binding to cpSRP43 >10-fold. Chaperone activities were measured and analyzed as in B and gave apparent Kd values of 1.9 µM, 168 nM, and 141 nM for cpSRP43(C175A), cpSRP43(Y287R_C175A), and cpSRP43(W299_C175A), respectively. Errors of Kd values were estimated to be ±10% (SD) based on at least two measurements (technical replicates).