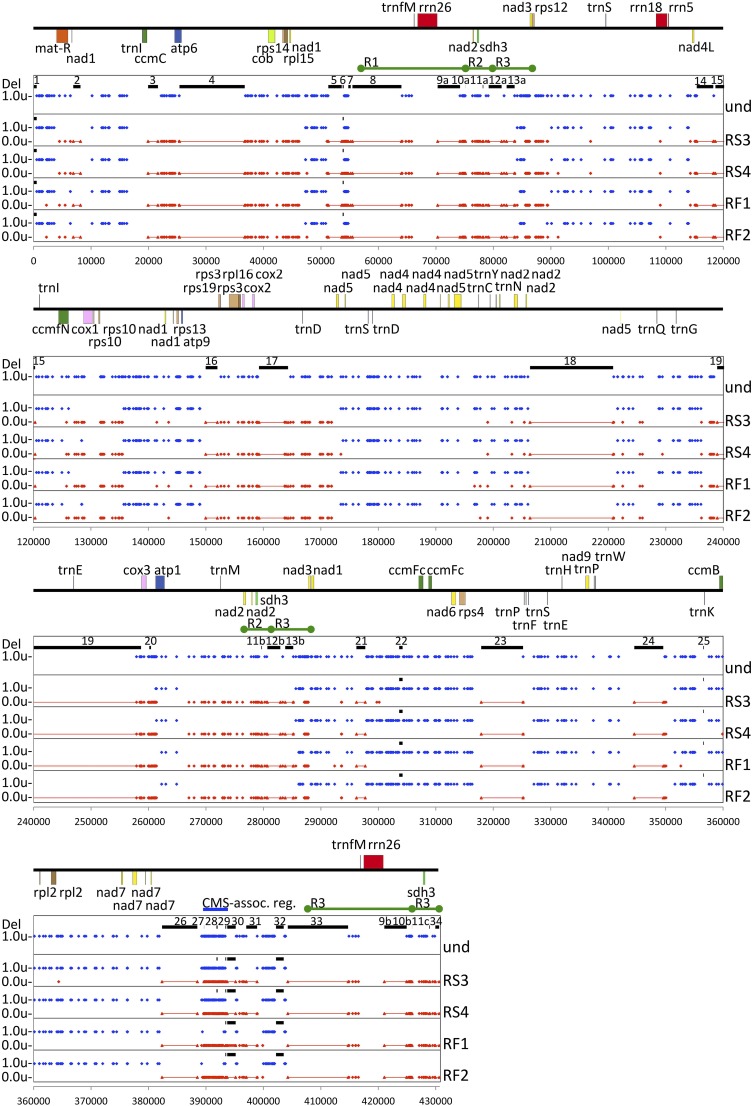
Fig. 3.
The mitochondrial genome of the GT19-C seed progeny is a mosaic of the two graft partners. We aligned the parent-specific SNPs in two fertile and two CMS recombinants using the N. sylvestris mtDNA as reference. Red and blue dots identify N. sylvestris- and N. undulata- specific SNPs, respectively. These data representations account for all N. sylvestris genes and exclude sequences unique to N. undulata (und). Deletions in the N. undulata mtDNA are shown as numbered black bars (SI Appendix, Table S5). N. sylvestris sequences absent in N. undulata and present in the recombinant progeny are red lines. The mitochondrial repeats are marked as R1, R2, and R3. The blue bar marks the CMS-associated region between nucleotides 389,706 and 393,005. The gene map was created using the Organellar Genome Draw program (49) based on the N. sylvestris mtDNA annotation (KT997964).