Fig. 3.
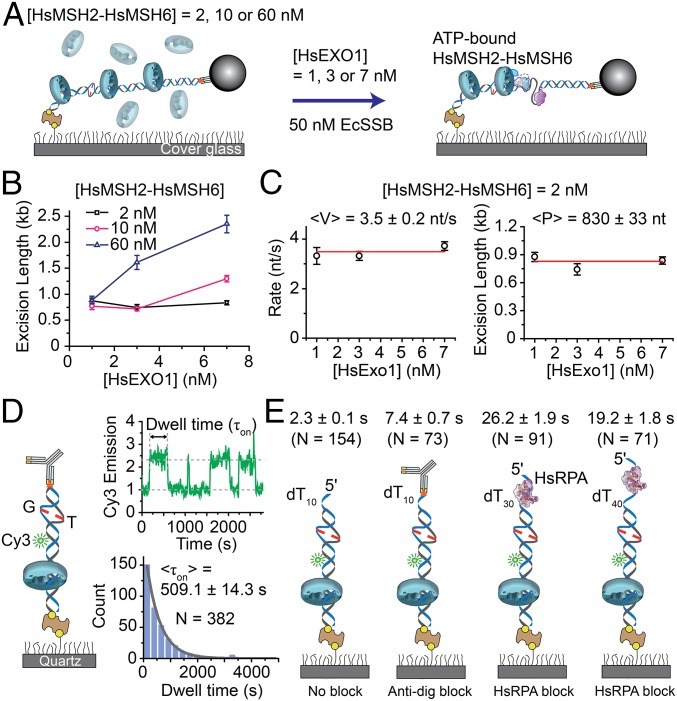
Multiple HsMSH2–HsMSH6 sliding clamps enhance HsEXOI excision. (A) Schematic illustration of the single molecule analysis of HsMSH2–HsMSH6 preassembled on mismatched DNA, followed by infusion of HsEXOI in the presence EcSSB. The HsMSH2–HsMSH6 concentration determines the number of sliding clamps loaded on the mismatched DNA (SI Appendix, Fig. S6). (B) The dependence of excision length (±SE) on HsEXOI concentration in the presence of different HsMSH2–HsMSH6 concentrations. The excision lengths were obtained from the analysis of ∼32–160 molecules. (C) The dependence of 5′-MMR excision rate (<V> ± SE) and processivity (<P > ± SE) on HsEXOI concentration in the presence of HsMSH2–HsMSH6 (2 nM) where 88% of the single mismatched DNA molecules contain a single sliding clamp. n = 32, 34, and 126 for 1 nM, 3 nM, and 7 nM HsEXOI. (D, Left) Schematic illustration of the smPIFE 40-bp mismatched DNA substrate (SI Appendix, Table S2). (Right Top) Representative time trace of HsMSH2–HsMSH6 lifetime (τon) analysis. (Right Bottom) Binned histogram fit with a single exponential decay to determine the lifetime (<τon> ± SE) of HsMSH2–HsMSH6 on tightly blocked-end mismatch DNA. (E) Average smPIFE lifetime (<τon>) for HsMSH2–HsMSH6 on various illustrated 40-bp mismatched DNA substrates (SI Appendix, Table S2). The DNA substrates used are (left to right): oligo-dT10 tail; oligo-dT10 tail containing 5′ dig-antidig blocked end; oligo-dT30 tail bound with HsRPA (20 nM); and oligo-dT40 tail bound with HsRPA (20 nM). Dwell times are shown above each illustration (±SE).