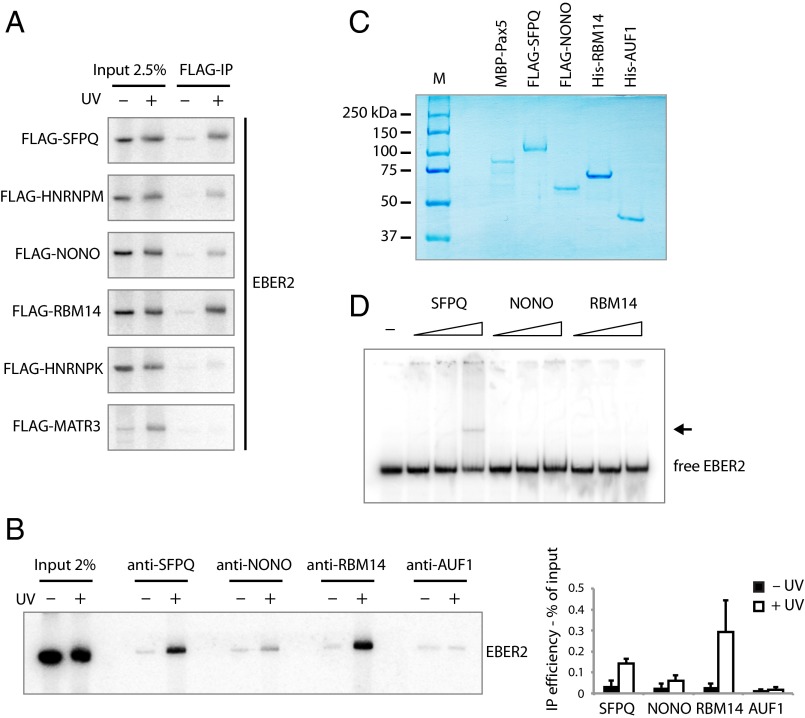
Fig. 2.
UV-crosslinking experiments and EMSAs identify proteins interacting with EBER2. (A) HEK 293T cells were transfected with plasmids expressing EBER2 and FLAG-tagged SFPQ, HNRNPM, NONO, RBM14, HNRNPK, or MATR3. EBER2 was expressed at levels comparable to those in BJAB-B1 cells (∼2.5 × 105 copies per cell); FLAG-tagged proteins were expressed at levels comparable to each other (Fig. S1B). Intact cells were irradiated with UV light where indicated. EBER2 was coimmunoprecipitated using anti-FLAG antibodies and detected by Northern blot analysis, showing direct interactions with SFPQ, HNRNPM, NONO, and RBM14. For the input lanes, RNA was isolated from 2.5% the amount of lysate used in the FLAG-IP. (B) Intact BJAB-B1 cells were irradiated with UV light where indicated and subjected to immunoprecipitation using antibodies against SFPQ, NONO, RBM14, and AUF1 as a negative control. EBER2 was detected by Northern blot analysis. EBER2 interacts directly with SFPQ, RBM14, and weakly with NONO. EBER2 quantification from three independent experiments (mean ± SD) is shown to the Right. (C) Coomassie-stained gel of recombinantly expressed proteins MBP–Pax5, FLAG–SFPQ, FLAG–NONO, His–RBM14, and His–AUF1p40 used for EMSA. SFPQ and NONO were purified from baculovirus-infected Sf9 cells, whereas the other proteins were purified from E. coli. (D) EMSA of EBER2 with SFPQ, NONO, and RBM14. EBER2 (1 nM in vitro transcribed) was incubated with increasing amounts of recombinant protein (5, 25, 125 molar excess) for 30 min on ice in EMSA buffer and resolved on a native polyacrylamide gel. An EBER2 bandshift is observed after incubation with recombinant SFPQ at the highest molar ratio (1:125). Free EBER2 is indicated; the arrow points to the EBER2–SFPQ complex.