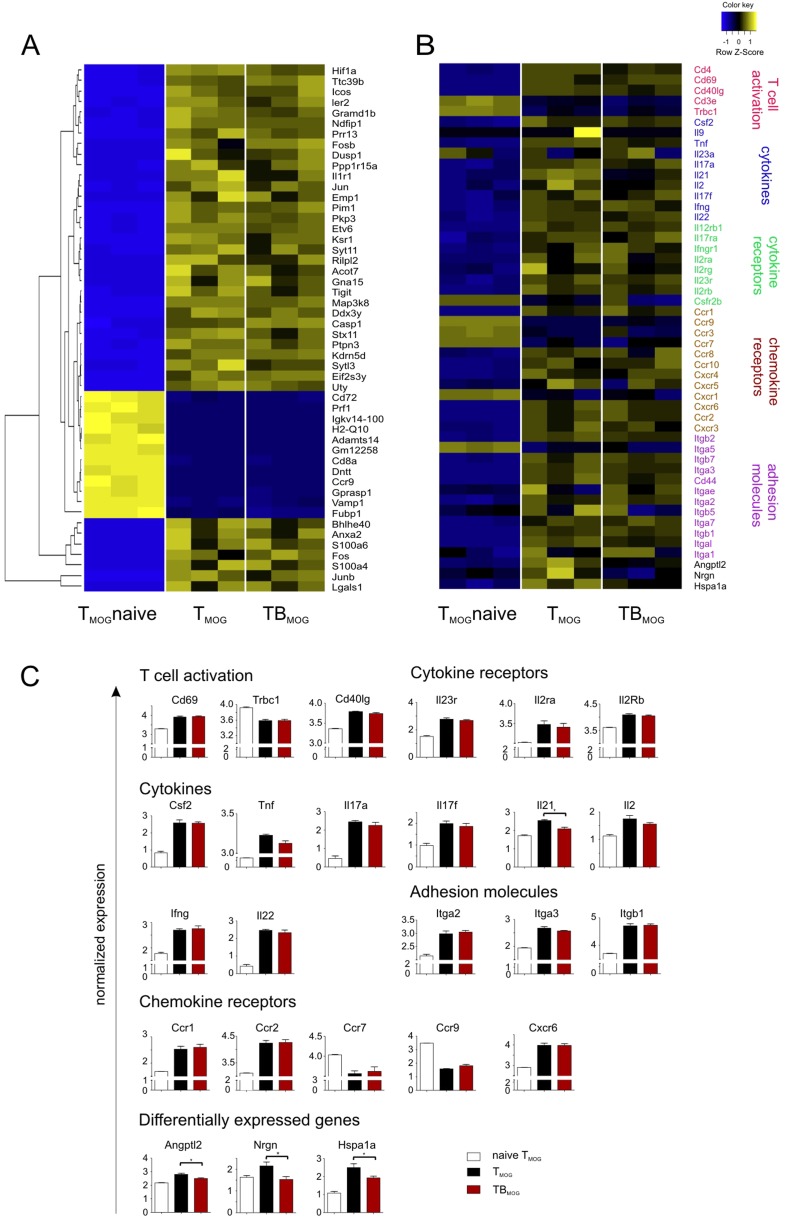
Fig. S4.
RNAseq transcriptome analyses of TMOG cells from T-MOG and T-/B-MOG mice and of naive TMOG cells. Transcriptomes of effector TMOG cells sorted from spleens of T-MOG and T-/B-MOG mice 9 d p.i. were compared and set in comparison with nonprimed TMOG cells (n = 3). (A) Heat-map of the top 50 most differentially expressed genes between naive TMOG cells and effector TMOG cells from T-MOG and T-/B-MOG mice. Individual values of three different biological replicates from naive TMOG cells and those from T-MOG and T-/B-MOG mice are shown. (B) Heat-map of 50 exemplary genes associated with relevant T-cell functions: i.e., activation and differentiation, cytokines and cytokine receptors, chemokine receptors, and cell locomotion genes. TMOG cells from nonimmunized animals and those from T-MOG mice and T-/B-MOG mice were compared. (C) Gene expression levels of individual genes were normalized and are depicted as mean ± SEM. Genes are grouped according to function comprising genes involved in T-cell activation, cytokines, cytokine receptors, chemokine receptors, and adhesion molecules. The last group lists the four genes (out of more than 38,000 genes) that showed significantly differential expression in TMOG cells from T-MOG mice and T-/B-MOG mice. Note that the differentially expressed Il-21 gene is grouped among the cytokine genes.