Fig. 6.
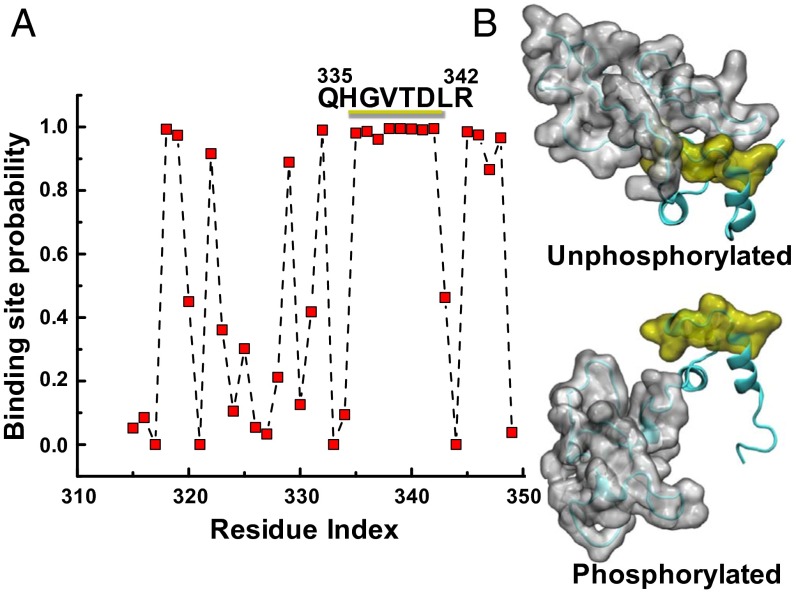
(A) A phosphorylated MyBP-C motif (residues 259–353) protein–protein binding site is predicted to be at residues 335–342 using cons-PPISP, in which a score of 0 is improbable and 1 is highly probable. See SI Appendix, SI Results and Table S3 for residue-specific values. (B) MD simulations suggest that this region is accessible for binding only when phosphorylated: A series of eight residues (yellow) is uncovered as the disordered N-motif (gray) shifts from the unphosphorylated (Upper) to the phosphorylated (Lower) conformation.