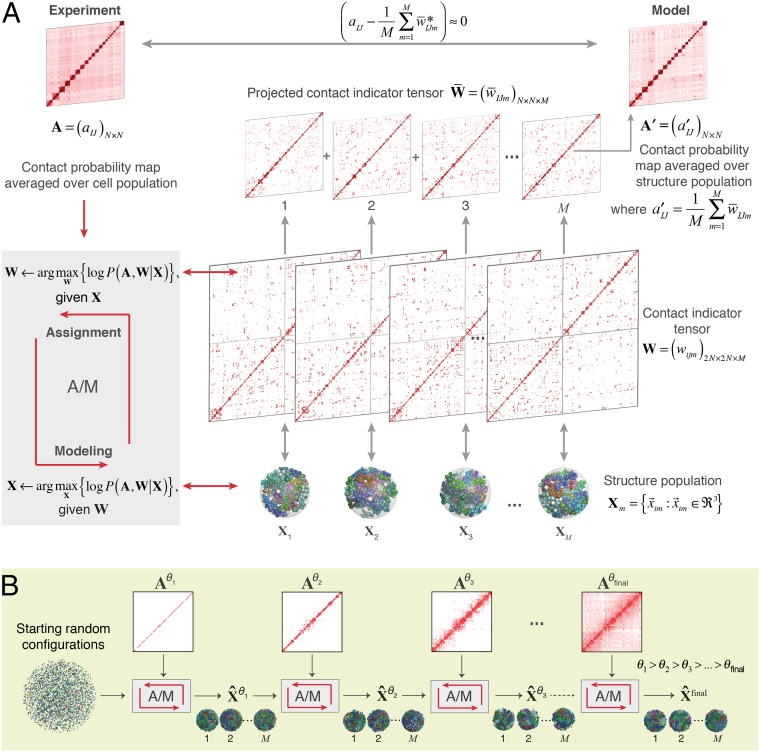
Fig. 1.
Schematic of the population-based genome structure modeling approach. (A) A population of M genome structures is constructed, in which the formation of contacts between chromosome domains over all structures is statistically consistent with the contact probability matrix A, derived from Hi-C experiments (Materials and Methods). We formulate this problem as a maximum likelihood estimation problem. Because the Hi-C data A are incomplete, we introduce the “contact indicator tensor” W, a binary third-order tensor that can complete the missing contact information in A. That is, W specifies which domain contacts exist in which structures of the population and also distinguishes between contacts from homologous chromosome copies. Also shown is the “projected contact indicator tensor,” , derived from W by projecting its diploid genome representation to its haploid representation (SI Appendix). (B) The maximum likelihood optimization is achieved through a stepwise iterative process, where we gradually increase the optimization hardness by gradually adding contacts of the matrix with decreasing contact probability threshold θ. This process generates a structure population that is consistent with the Hi-C data (SI Appendix).