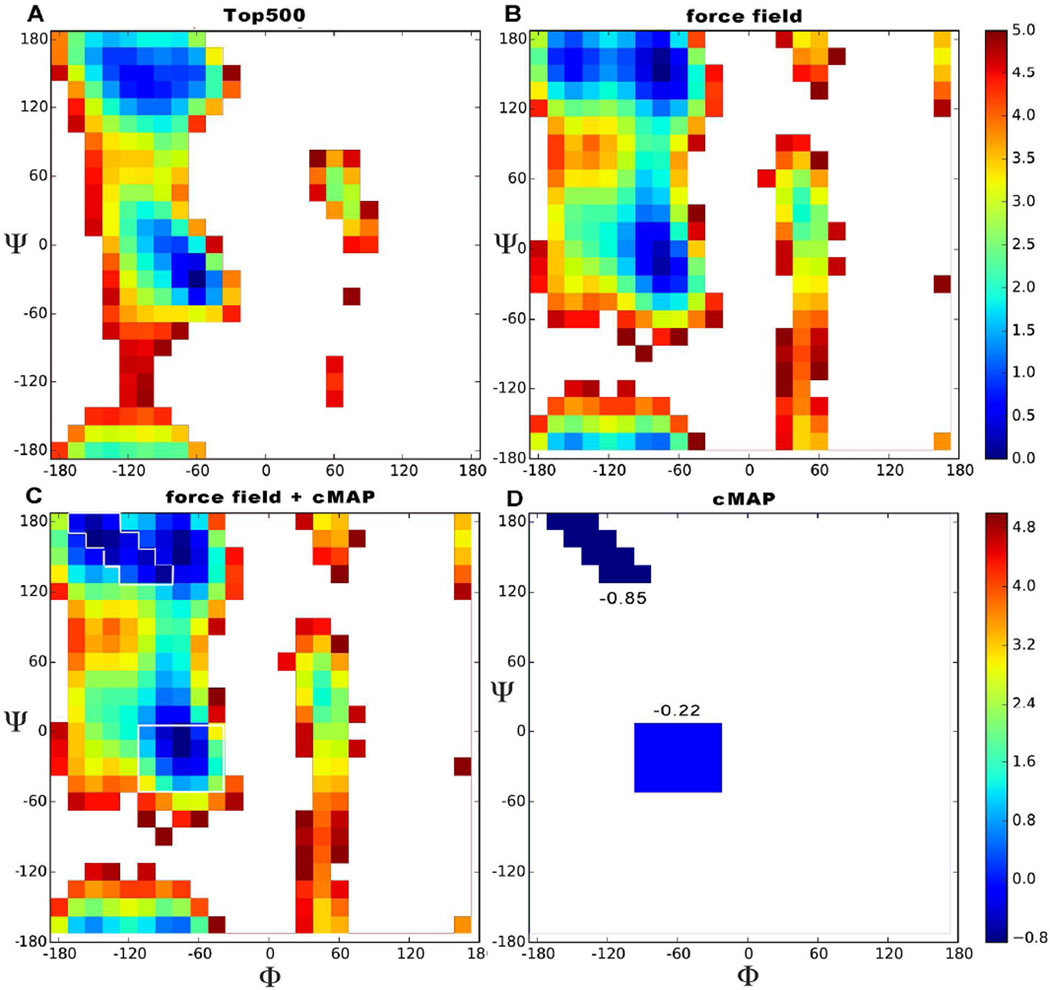
Figure 1.
Free energy ϕ,ψ map. The color scale indicates relative energies with respect to the lowest energy conformation. Energies are truncated at 5 kcal/mol. The experimental plot (A) was derived from data from the top500 dataset by looking at all the non glycine, proline or pre-proline residues, binning them in a 2D histogram, counting populations and using the Boltzmann relation to estimate free energies. The calculated plot (B) was derived from simulations of trialanine (ff12SB force field + Gbneck2 implicit solvent). Plot C shows the effect of ff12SB-cMAP on top of ff12SB for Alanine; the regions directly influenced by cMAP are outlined in white for clarity. The cMAP energy correction we apply is shown by itself in panel D.