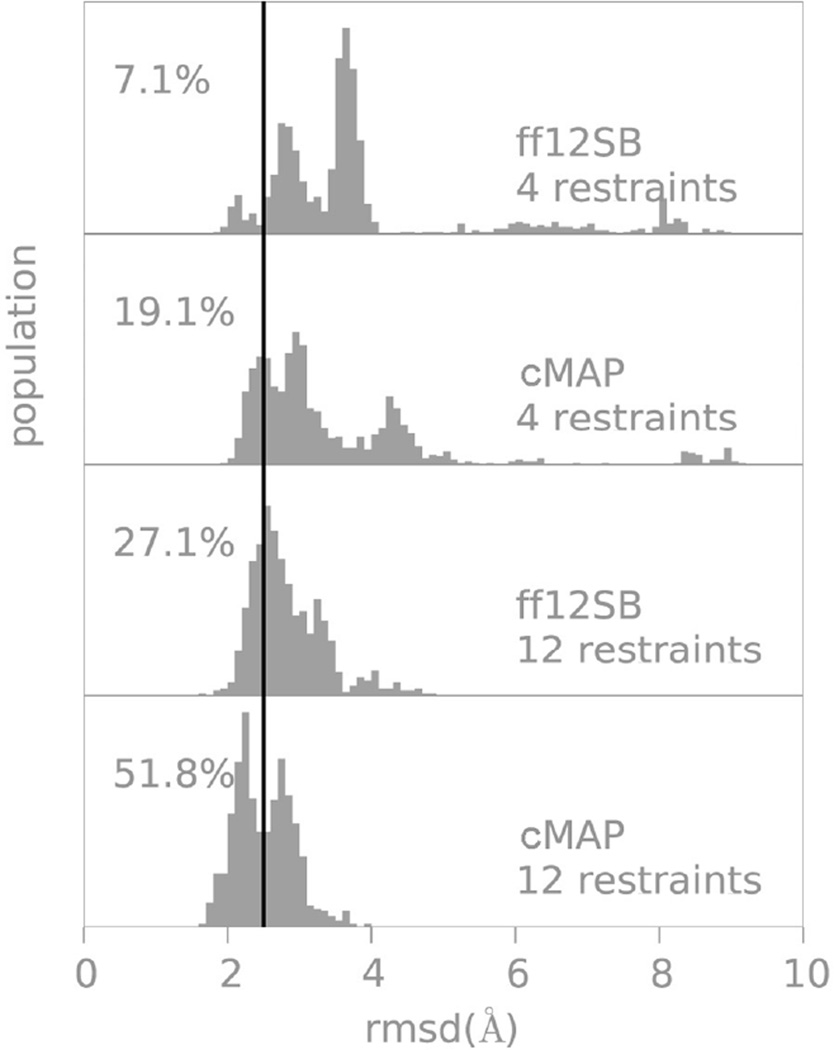
Figure 3.
RMSD distributions from native in protein G folding simulations with different force fields and number of restraints. The percentages refer to the RMSD populations below 2.5Å sampled in the last 50 ns of a 60 ns restrained folding trajectory. We use a stringent value of 2.5Å as a cutoff since the use of restraints already favor a native like topology; notice that in all cases most conformations are within 4Å from the experimental structure.