FIGURE 2.
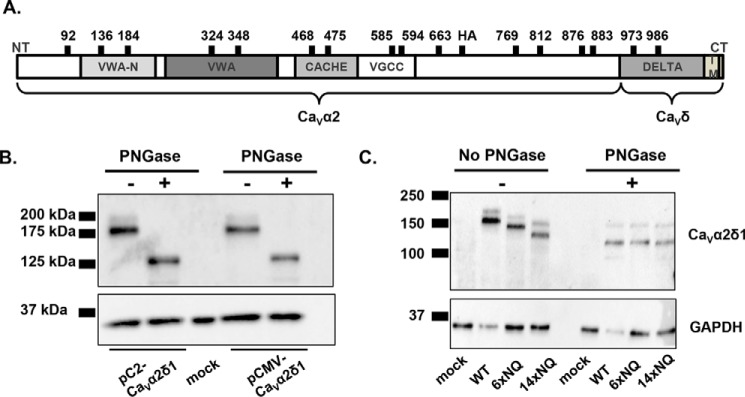
PNGase F-mediated deglycosylation of the N-linked sugars from CaVα2δ1 expressed in HEKT. A, relative positions of the predicted N-glycosylation sites are shown on the structural domains of the rat CaVα2δ1. Structural domains were identified using protein BLAST (blast.ncbi.nlm.nih.gov) with the UniProtKB/Swiss-Prot database. The four structural domains are shown by boxes: VWAN (NCBI pfam08399), VWA (NCBI smart00327), CACHE (NCBI pfam02743), and VGCC (NCBI pfam08473). B, cells were transiently transfected with pCMV-CaVα2δ1 or pC2-CaVα2δ1. Total cell lysates were extracted 24 h after transfection using the protocol described earlier. Total cell lysates were denatured 10 min at 95 °C before incubation in the absence (−) or presence (+) of PNGase during 1 h at 37 °C. Proteins were electrophoresed on a 8% SDS-polyacrylamide denaturating gel, transferred to a nitrocellulose membrane, and probed with an anti-CaVα2δ1 (Alomone Labs) and anti-GAPDH as a loading control. Each lane was loaded with 10 μg of proteins. As seen, the electrophoretic mobility of the CaVα2δ1 protein decreased by 50 kDa following enzymatic digestion. C, mutations of multiple glycosylation sites decreased protein mobility. HEKT cells were transiently transfected with pmCherry-CaVα2δ1-HA WT, 6xNQ, or 14xNQ. Exactly 24 h after transfection, cells were lysed, and protein lysates were either treated with the vehicle buffer or with PNGase F during 1 h at 37 °C. Proteins were fractionated by SDS-PAGE (8%). Western blot analysis was carried out with the CaVα2δ1 antibody (Alomone Labs) as the primary antibody, and signal was detected using the Bio-Rad ECL substrate. The 2nd lanes (± PNGase) were loaded with 5 μg of proteins, and the 1st, 3rd, and 4th lanes were loaded with 10 μg. 1st lane, mock-transfected HEKT cells; 2nd lane, pmCherry-CaVα2δ1-HA WT; 3rd lane, pmCherry-CaVα2δ1-HA 6xNQ; 4th lane, pmCherry-CaVα2δ1-HA 14xNQ. The calculated molecular masses for the high density band are before treatment with PNGase as follows: 2nd lane, 171 kDa; 3rd lane, 155 kDa; 4th lane, 133 kDa; after digestion with PNGase: 2nd lane, 123 kDa; 3rd lane, 123 kDa; and 4th lane, 123 kDa. The 10-kDa difference in the molecular masses between the 14xNQ before and after treatment with PNGase could suggest that N-glycosylation was not completely eliminated in the 14xNQ mutant or else that the enzymatic treatment itself altered the migration of the protein in the gel.