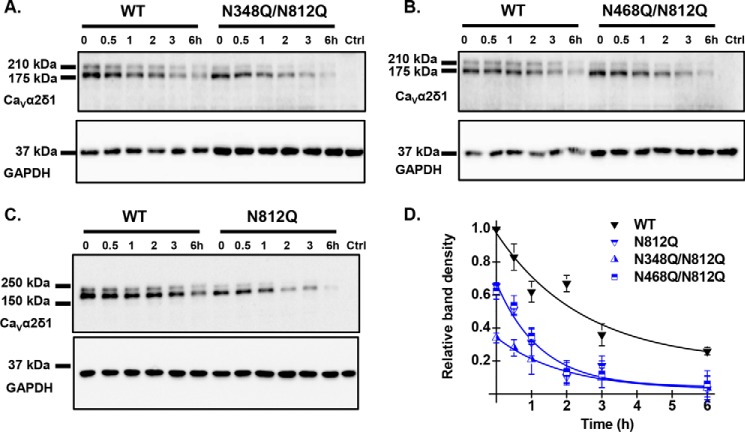
FIGURE 9.
Multiple Asn mutations may impair protein stability. HEKT cells were transiently transfected with pmCherry-CaVα2δ1-HA WT and the glycosylation mutants N348Q/N812Q (A), N468Q/N812Q (B), and N812Q (C). Exactly 24 h after transfection, subconfluent cells were incubated with 100 μg/ml cycloheximide. At the indicated time points, cell lysates were prepared and fractionated by SDS-PAGE (8%). CaVα2δ1 and GAPDH proteins were respectively probed with anti-CaVα2δ1 (Alomone Labs) and anti-GAPDH. Please note that we loaded 2× more proteins in the wells for the double mutants to visualize their time course alongside the WT construct. In particular, each lane for CaVα2δ1-HA WT was loaded with 5 μg of proteins in A and B, and 10 μg of proteins were loaded for N348Q/N812Q and N468Q/N812Q. C, 10 μg of proteins were loaded for CaVα2δ1-HA WT and N812Q. D, protein density was estimated relative to the density of the GAPDH band and normalized to the protein density for the wild-type construct at time 0. Protein band intensities were quantified by densitometry using ImageLab (Bio-Rad) software at a single exposure selected for clear bands without saturation. Averaged data points were fitted with a mono-exponential decay function. The half-life was t½ = 2.8 ± 0.5 h (n = 7) for pmCherry-CaVα2δ1-HA WT; t½ = 1.2 ± 0.8 h (n = 2) for N348Q/N812Q; t½ = 1.8 ± 0.5 h (n = 3) for N468Q/N812Q; and t½ = 1.9 ± 0.7 h (n = 3) for N812Q. As seen, the data points for the decay of N812Q and N468Q/N812Q are superimposed with similar relative densities, whereas protein density for N348Q/N812Q was lower at time 0.