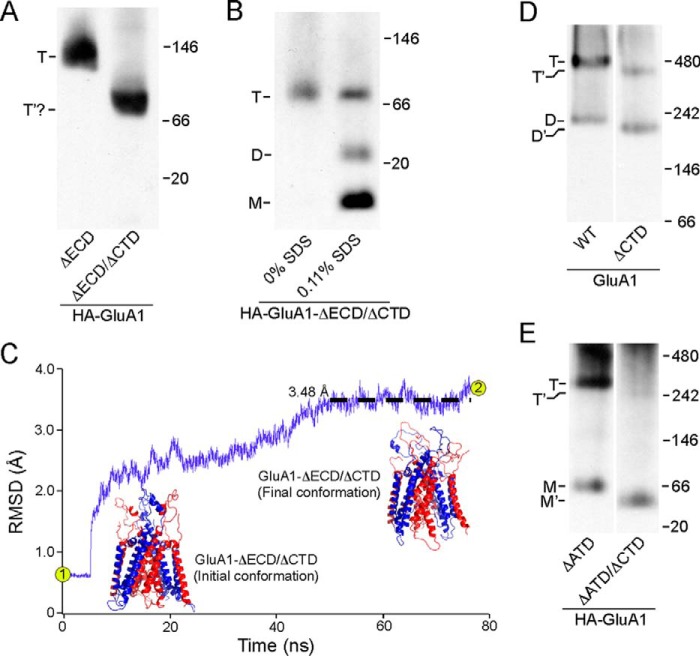
FIGURE 3.
Tetramerization of GluA1 with or without the CTD. A, BN-PAGE of HA-GluA1-ΔECD as well as the HA-GluA1-ΔECD/ΔCTD. For HA-GluA1-ΔECD/ΔCTD, a single band was detected (indicated as T′?) with an estimated molecular mass roughly consistent with that of a tetramer (91.6 kDa). B, BN-PAGE of HA-GluA1-ΔECD/ΔCTD harvested either without SDS or with 0.11% SDS in the solubilization buffer. C, homology modeling and MD simulation of an ECD/CTD-free GluA1 tetramer based on 3KG2 (see “Experimental Procedures”). R.m.s. deviation between the model and corresponding regions of Protein Data Bank 3KG2 over time is shown in purple (black dashed line shows averaged r.m.s. deviation between 50 and 76.7 ns). Initial conformation of the model after energy minimization (point 1 in r.m.s. deviation trace) was almost identical to 3KG2, whereas the final conformation after 76.7 ns of MD simulation (point 2 in trace) showed minimal changes (r.m.s. deviation <3.5 Å). D and E, BN-PAGE of GluA1 and GluA1-ΔCTD (D) or HA-GluA1-ΔATD and HA-GluA1-ΔATD/ΔCTD (E). Estimated oligomeric states are indicated as described in the legend to Fig. 2.