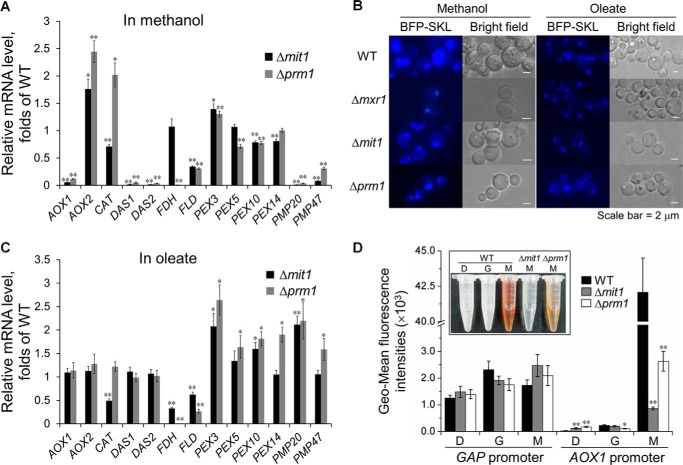
FIGURE 2.
Mit1 activates methanol utilization genes but keeps away from peroxisome functions. A and C, transcription levels of genes involved in methanol utilization pathway and peroxisome biogenesis of the Δmit1 and Δprm1 strains grown in methanol (A) or oleate (C). The mRNA levels were normalized to the housekeeping gene ACT1 in each sample. The relative expression level indicated on the y axis (2−ΔΔCT) for each gene was normalized to that in the WT cells grown in the corresponding carbon source. The error bars represent the standard deviation of three biological replicates, each with three technical replicates, assayed in duplicate. B, fluorescence microscopy images of Δmit1, Δprm1, Δmxr1, and WT strains expressing the peroxisome-targeted BFP-SKL fusion protein in methanol and oleate. SKL was fused to the C terminus of BFP to localize BFP to the peroxisomes. The Δmit1, Δprm1, Δmxr1, and WT cells were pregrown in YPD to log phase and washed three times in sterile water. The washed cell pellets were transferred to YNM medium (YNB + 0.5% methanol) and YNO medium (YNB + 0.5% oleate + 0.05% Tween 80) supplemented with the requisite amino acid. After culture at 30 °C for 1 h, aliquots of cell pellets were harvested and subsequently washed to visualize BFP-SKL. D, evaluation of the activity of PAOX1 by colorable reaction of Aox and exploiting a reporter gene (GFP) expression assay in the Δmit1, Δprm1, and WT strains grown in glucose, glycerol, and methanol, respectively. For the GFP expression assay, the Δprm1 strain was used as a positive control. GFP expressed under the control of PGAP was used as a negative control. GFP expression in P. pastoris was analyzed using an enzyme-labeled instrument (BioTek Instruments) at an excitation wavelength of 488 nm and an emission wavelength of 512 nm. The fluorescence values were determined by a geometric mean (Geo-Mean) method. The error bars represent the standard deviation of three biological replicates, each with three technical replicates, assayed in duplicate. An independent-sample t test was used to determine the statistical significance of the mutant groups relative to the WT groups in the corresponding carbon sources and promoter. *, p < 0.05; ** p < 0.01. Colorable reaction was performed as per Zhang et al. (9). It was visualized by adding the Aox reaction mixture with the permeabilizing agent to the cell pellets for 30 min. The higher Aox activity corresponds to the deeper red of the reaction mixture. D, glucose; G, glycerol; M, methanol.