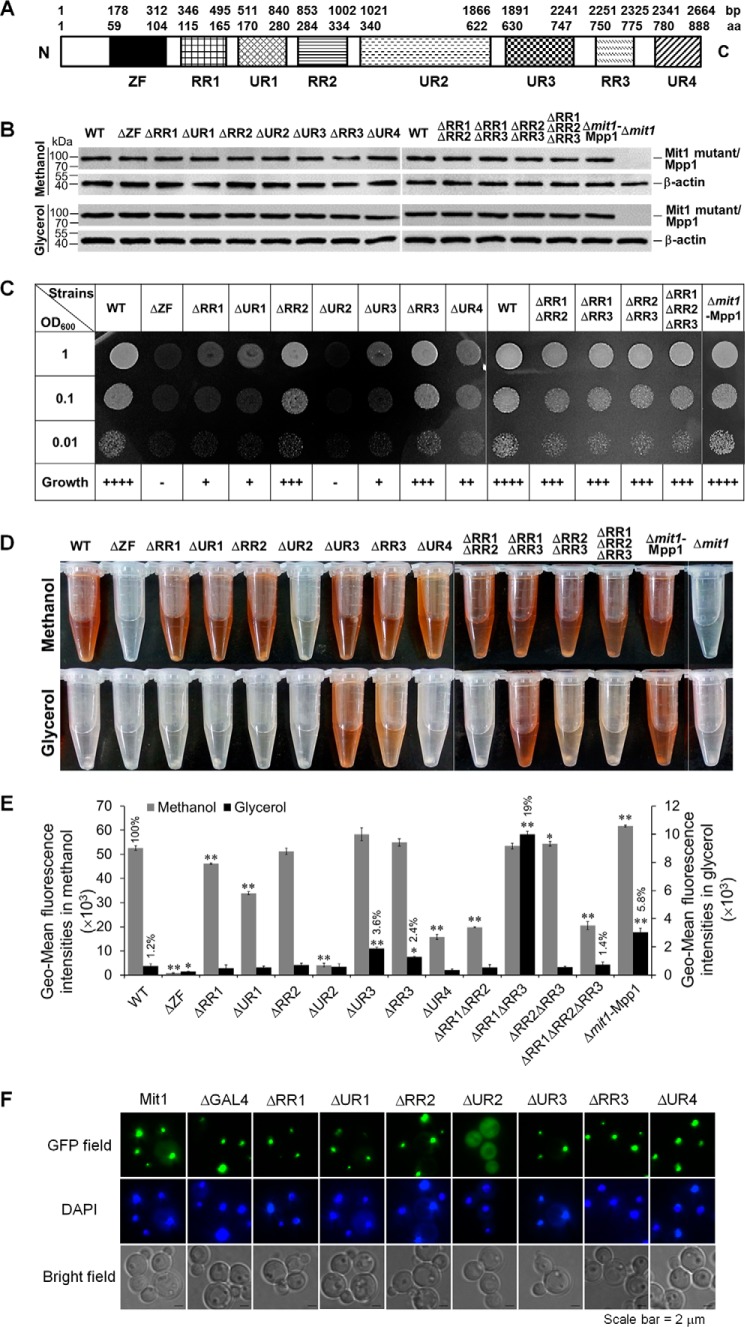
FIGURE 3.
Complementation of the P. pastoris Δmit1 mutant by H. polymorpha Mpp1 and domain-deleted Mit1. A, schematic drawing of the annotated domains within Mit1. ZF, the GAL4-like Zn(II)2Cys6 (or C6 zinc) binuclear cluster DNA-binding domain. B, expression of Mpp1 and domain-deleted Mit1 in complementary cells in glycerol and methanol. Mpp1 and domain-deleted Mit1 were FLAG-tagged at the C terminus and detected by Western blotting using anti-FLAG antibody. Twenty micrograms of total protein was loaded into each lane, and the expression of β-actin was used as a positive control. Normalization of the signal intensity to total protein loading is described under “Miscellaneous Methods.” C, growth of Δmit1-Mpp1 and domain deletion mutants of Mit1 in methanol. Cells were pregrown in YPD medium to log phase and washed three times in sterile water. The washed cell pellets were then diluted to an OD600 of 0.01, 0.1, and 1 (as indicated). Ten μl of each was spotted onto YNB plates containing the 0.5% methanol and the requisite histidine. Then the plates were incubated at 30 °C for about 3 days. ++++, the same growth as WT; +++, little growth defect compared with WT; ++, weak growth; +, little growth; −, no growth. D, colorable reaction of Aox in Δmit1-Mpp1 and domain deletion mutants of Mit1 in methanol and glycerol. A colorable reaction was performed the same as described for Fig. 2. E, evaluation of the activity of PAOX1 by exploiting a reporter gene (GFP) expression assay in Δmit1-Mpp1, domain deletion mutants of Mit1, and the WT grown in methanol and glycerol. The GFP expression was measured the same as described for Fig. 2. Δ, strains with domain-deleted Mit1. The error bars represent the standard deviation of three biological replicates, each with three technical replicates, assayed in duplicate. An independent sample t test was used to determine the statistical significance of the mutant groups relative to WT groups in corresponding carbon sources. *, p < 0.05; **, p < 0.01. F, subcellular localization of Mit1 and domain-deleted Mit1 with GFP fusing at their N termini in methanol media. DAPI was used to stain the cell nucleus.