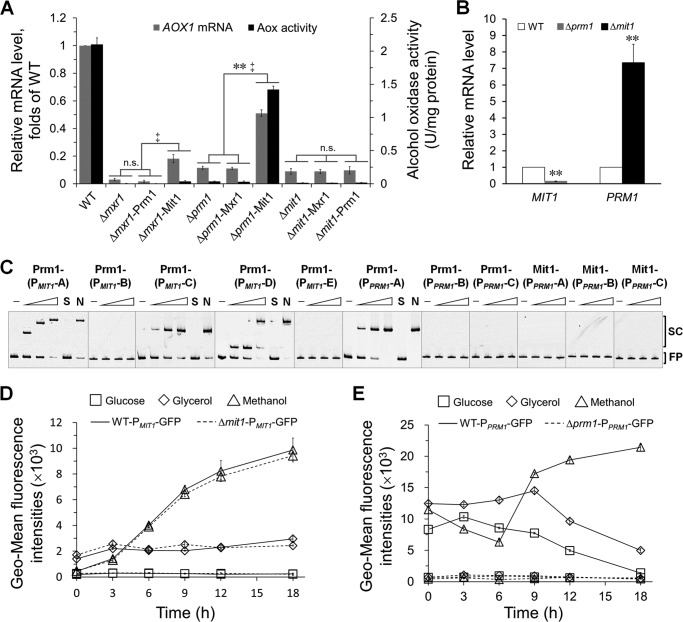
FIGURE 7.
Derepression/induction of AOX1 promoter by Mit1, Prm1, and Mxr1. A, relative mRNA levels of AOX1 and enzyme activity of Aox in mutant strains. The mRNA levels were normalized to that of the housekeeping gene ACT1 in each sample. The relative expression level indicated on the y axis (2−CT) for each gene was normalized to that in the WT strain grown in methanol. The enzyme activity of Aox was assayed as described previously (37, 38). A unit of Aox represents 1 μmol of product min−1 mg−1 of protein at 30 °C. The error bars represent the standard deviation of three biological replicates, each with three technical replicates, assayed in duplicate. An independent-sample t test was used to determine the statistical significance of different groups as indicated. AOX1 mRNA: **, p < 0.01. Aox activity: ‡, p < 0.05; n.s., no significance. B, detection of mRNA levels of PRM1 in the Δmit1 mutant and MIT1 in the Δprm1 mutant in methanol. Other labeling is the same as in A. An independent-sample t test was used to determine the statistical significance of mutant groups relative to WT groups for corresponding gene. **, p < 0.01. C, EMSAs of P. pastoris Prm1 or Mit1 with fragments of the MIT1 promoter (PMIT1) and the PRM1 promoter (PPRM1). EMSAs were performed by incubating Cy5-labeled PMIT1-A (−1,000 to −701 bp of MIT1), PMIT1-B (−800 to −601 bp of MIT1), PMIT1-C (−700 to −351 bp of MIT1), PMIT1-D (−450 to −251 bp of MIT1), PMIT1-E (−350 to −1 bp of MIT1), PPRM1-A (−699 to −540 bp of PRM1), PPRM1-B (−562 to −314 bp of PRM1), and PPRM1-C (−338 to −112 bp of PRM1) with the zinc finger domains of P. pastoris Prm1 (aa 41–90) or Mit1 (aa 1–150) expressed by E. coli. The experimental procedure and other labeling were the same as described in the legend for Fig. 4A. D and E, detection of self-regulation of Mit1 (D) and Prm1 (E). GFP expressed by PMIT1 and PPRM1 was used to quantify the self-regulation of Mit1 and Prm1 in the strains of WT-PMIT1-GFP, Δmit1-PMIT1-GFP, WT-PPRM1-GFP, and Δprm1-PPRM1-GFP. GFP expression was analyzed as described in the legend for Fig. 2.