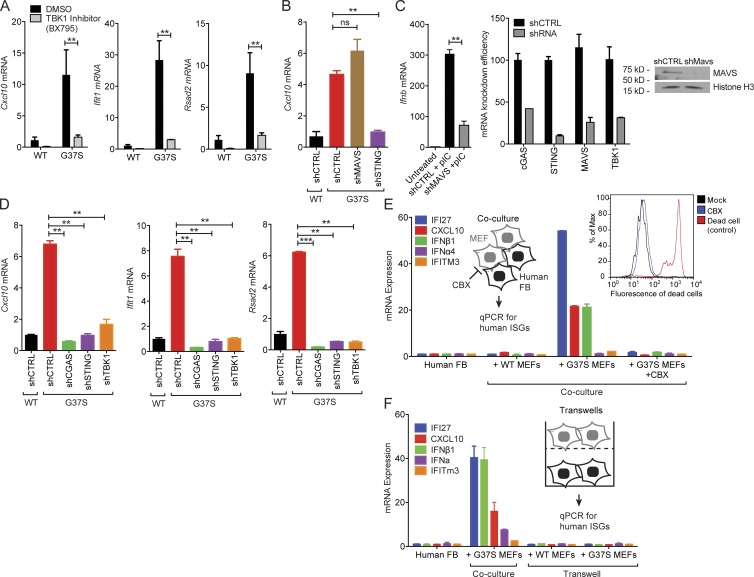
Figure 2.
Immune activation in Rnaseh2aG37S/G37S primary MEFs requires the cGAS–STING innate immune pathway. (A) Quantitative RT-PCR analysis of Cxcl10, Ifit1 and Rsad2 mRNA (all ISGs) in WT and Rnaseh2aG37S/G37S (G37S, same below) MEFs treated with DMSO or TBK1 inhibitor BX795 (10 µM) for 6 h. (B) Quantitative RT-PCR analysis of Cxcl10 mRNA in WT and G37S MEFs treated with shRNA against indicated genes involved in cytosolic nucleic acid-sensing. (C) shMAVS knockdown reduces poly(I:C)-induced IFN response. Knockdown efficiency is shown on the right. (D) Quantitative RT-PCR analysis of Cxcl10, Ifit1, and Rsad2 mRNA in WT and G37S MEFs treated with shRNA against indicated genes involved in DNA-sensing pathway. (E) Quantitative RT-PCR analysis of a panel of human ISGs and IFN genes in human fibroblasts (BJ-1 cells) co-cultured with WT or G37S MEFs for 18 h, with or without CBX treatment (inhibits gap junction). Left inset shows a schematic diagram of the gap junction assay. Right inset shows FACS analysis of cell death in mock- and CBX-treated cells. (F) Quantitative RT-PCR analysis of human ISGs in human fibroblasts in a trans-well assay co-cultured with WT or G37S MEFs for 18 h. Mice were compared with littermate controls. **, P < 0.01; ***, P < 0.001. ns, not significant. Data are representative of at least three independent experiments. Error bars represent the SEM. Unpaired Student’s t test (A–D).