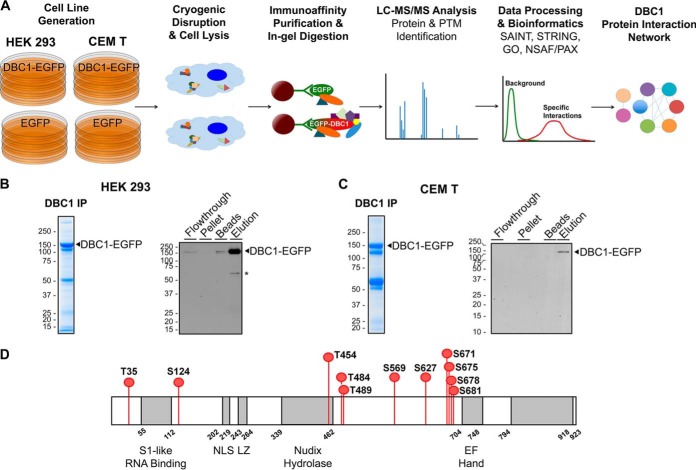
Fig. 3.
Workflow for investigating DBC1 protein interactions and phosphorylation sites. A, Proteomic workflow for immunoaffinity purifications (IP) of DBC1-EGFP and EGFP from stable CEM T and HEK293 cell lines. Immunoisolates were subjected to label-free IP-MS analysis with interaction specificity determined by SAINT. DBC1 protein interaction networks were constructed and specific DBC1 interactions were functionally clustered (using GO annotations, the UniProt database, and literature searches). B–C, Representative Coomassie-stained gel lanes of DBC1-EGFP isolations, and assessment of the isolation efficiency by Western blotting (10% of each fraction was loaded). B, CEM T DBC1-EGFP cell line and C, HEK 293 DBC1-EGFP cell line. *, nonspecific band. D, DBC1 phoshorylation sites identified in this study following DBC1-EGFP isolation from both CEM T and HEK293 cells.