Fig. 10.
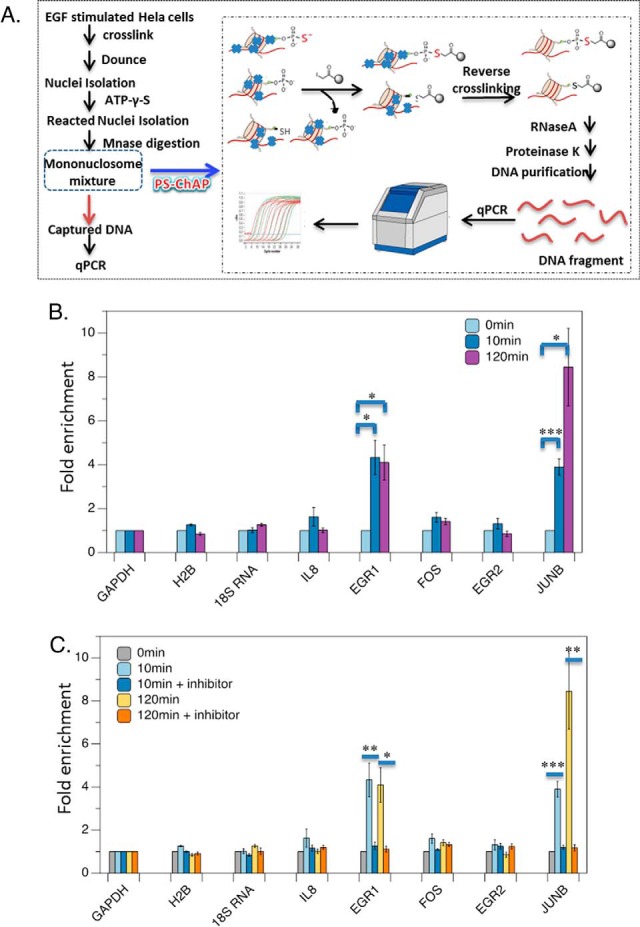
PS-ChAP for monitoring gene expression linked to cellular signaling. (A) EGF stimulated cells are crosslinked and then dounced to get the nuclear pellet followed by treating with ATP-γ-S. Illustration of the steps in the PS-ChAP strategy performed on a mononucleosome mixture after MNase digestion. By using iodoacetyl-beads, thiophosphorylated mononucleosomes can be isolated. Afterward, crosslinks were reversed and PS-ChAP captured DNA was purified and amplified by qPCR. (B) HeLa cells were serum starved for 24 h in media without serum and then treated with 100 ng/ml EGF for 10 min or 120min. Cells were harvested, and DNA was purified and analyzed by qPCR as shown in Fig. 10A. Statistically significant changes are indicated as ***p < .005, **p < .02 and *p < .05. (C) HeLa cells were serum starved for 24 h in media without serum, pretreated with 10 μm PD98059 for 1 h and then treated with 100 ng/ml EGF for 10min or 120min. Cells were harvested, and DNA was purified and analyzed by qPCR as shown in Fig. 10A. Values are relative to MEK-inhibitor-treated cells. Statistically significant changes are indicated as ***p < .005, **p < .02 and *p < .05. p values were calculated using the two-tailed unpaired Student's t test with equal variances. All error bars represent s.d. Data in parts B and C are the result of triplicate independent biological experiments.
