Fig. 3.
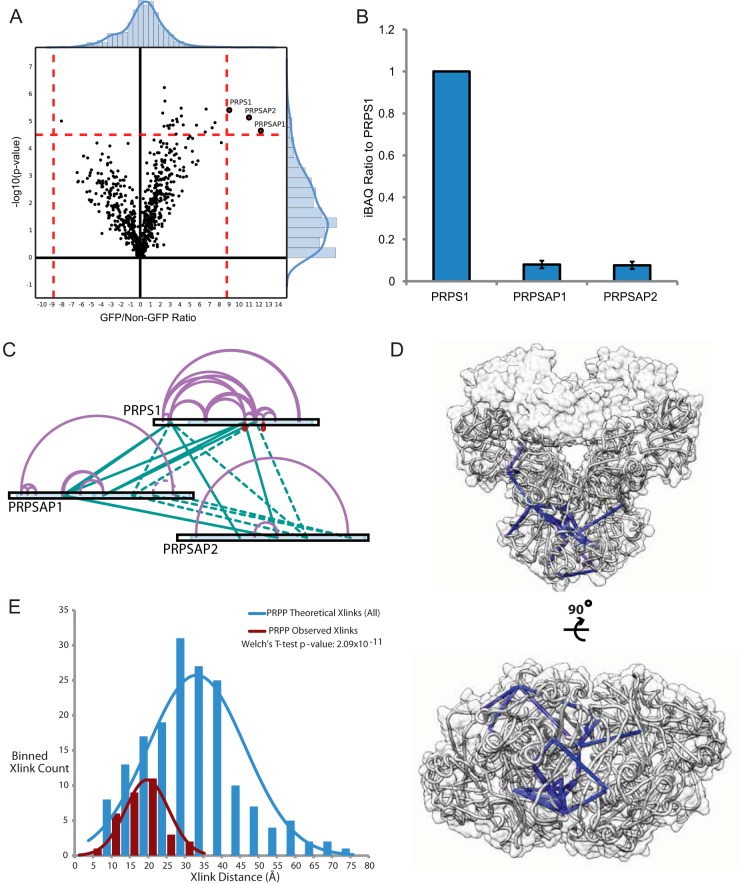
xIP-MS analysis of PRPP reveals high confidence distance constraints. A, LFQ analysis of PRPS1 was performed with triplicate pulldowns. Outliers are indicated in red, and background binders are indicated in black. Outlier cutoffs were drawn such that no proteins were present in the non-GFP quadrant on the volcano plot. B, Stoichiometry analysis was performed as described. All ratios are calculated by setting the bait (PRPS1) equal to 1. Error bars indicate standard deviations from triplicate samples. C, Identified cross-links for PRPP. The cross-link map was drawn in xiNet. Purple lines indicate self-links, and green lines indicate interprotein cross-links. Ambiguous cross-links (the same peptide in multiple proteins) are indicated by dashed lines. Homotypic cross-links (the same peptide cross-linked to itself, indicating multimerization) are drawn in red. Uniprot annotated domains are colored variously along the protein sequence bar. D, PRPS1 cross-links mapped onto the PDB: 2h06 crystal structure. Cross-links under 34 Å are colored in blue. PRPS1 monomers are colored in light gray. The silhouette of the PRPS1 holo-hexamer surface is shown in transparent gray.