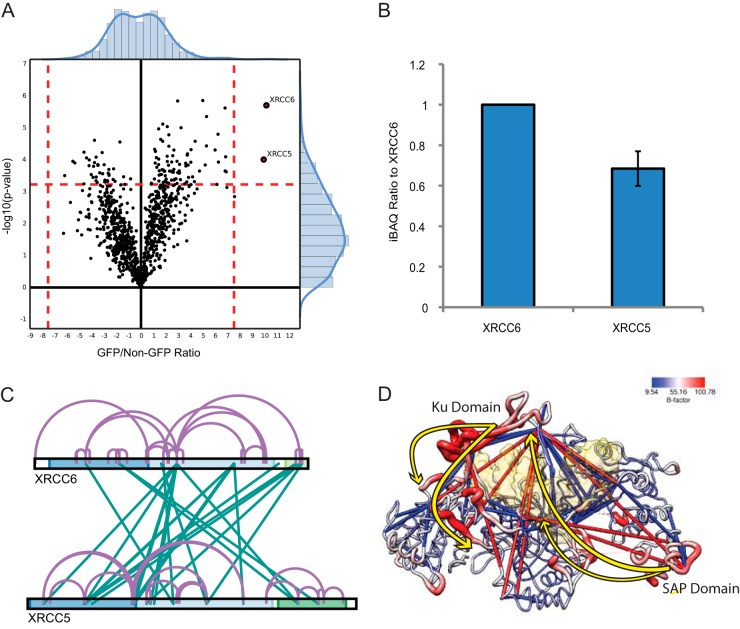
Fig. 5.
xIP-MS reveals conformational changes in XRCC5/6. A, LFQ identifies XRCC5/6 as robust interactors, in agreement with our gel analysis. B, XRCC5/6 stoichiometry is nearly 1:1 as indicated by iBAQ values. C, Cross-linking within the XRCC5/6 heterodimer. The cross-link map is colored as described previously. D, XRCC5/6 are colored by B-factor as indicated on the key; the protein backbone radii are scaled similarly by size. Cross-links within 40 Å are shown in blue, and cross-links longer than this threshold are shown in red. Major conformational shifts explaining observed cross-linking patterns are displayed as yellow arrows. The structure of PDB: 1jeq is used to display cross-links as the SAP domain is resolved only in this structure; however, the surface of the DNA structure bound to XRCC5/6 from PDB: 1jey is displayed in transparent yellow, after alignment with 1jeq using MatchMaker in UCSF Chimera.