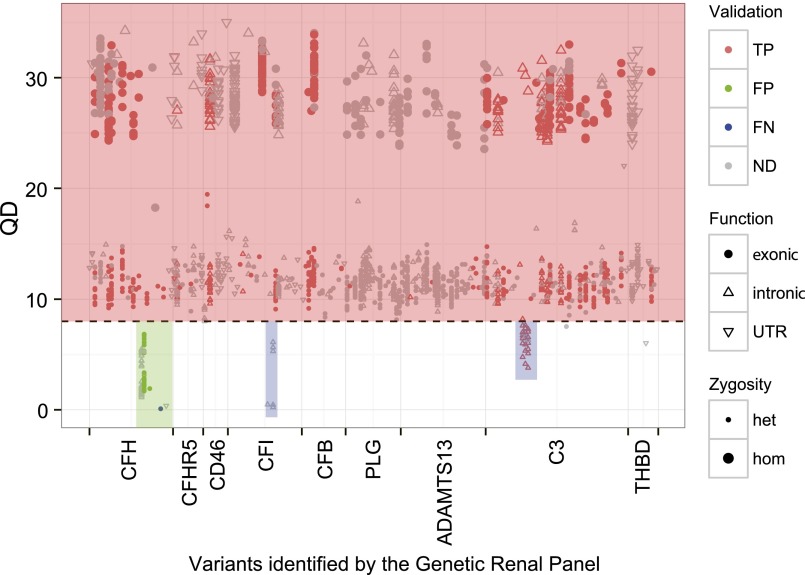
Figure 2.
Ninety-two subjects were analyzed using both TGE&NGS and Sanger sequencing as a validation step for the GRP. Nearly all exonic variants have very high quality (red box), reflecting an effective enrichment strategy. The single exception was a small portion of CFH (green box), where we observed both false-positive and false-negative calls. For this reason, exons 20–22 of CFH are always Sanger-sequenced (see Supplemental Figure 1). Some low QD variants in introns of CFI and C3 (blue box) were ignored because they did not impact exons and splice sites. TP, true positive; FP, false positive; FN, false negative; ND, no Sanger sequencing data; UTR, untranslated region.