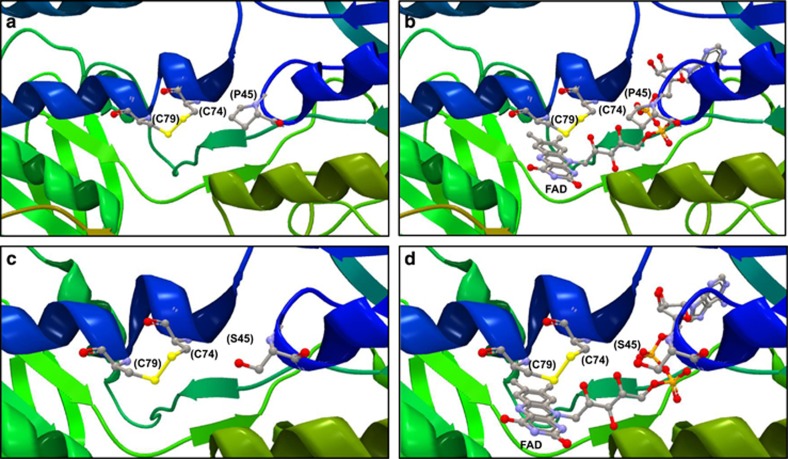
Figure 2.
Structure of the DLD and glutathione reductase protein-active sites. The protein structures have not been determined for either T. castaneum or R. dominica; therefore, the highly conserved orthologues of the DLD and glutathione reductase proteins from Saccharomyces cerevisiae (1JEH and 2HQM; Brautigam et al., 2005; Yu and Zhou, 2007) are shown to illustrate the location of the P>S variant amino acids relative to the active sites of the respective proteins. The extreme degree of sequence conservation between the paralogous enzymes is reflected in easily recognisable structural similarity between the DLD protein (a, b) and glutathione reductase (c, d). The placement of the FAD cofactor shown in b, d indicates that the P>S change is unlikely to affect the electron transfer reaction that occurs between the cysteine residues and the three-ring structure of the FAD. The cysteine (as the disulfide) and proline residues are highlighted and numbered according to the equivalent T. castaneum coordinates.