FIGURE 1.
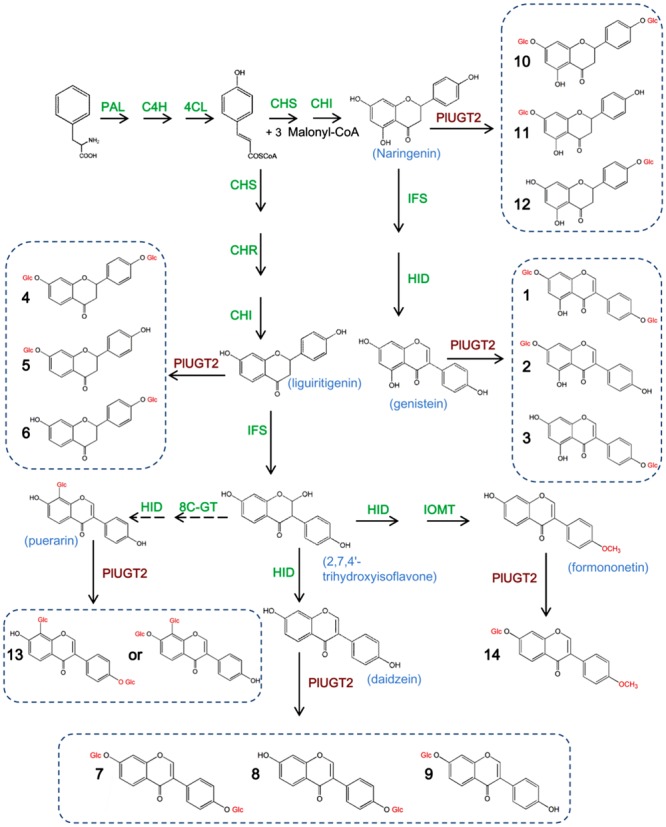
Proposed pathways for isoflavone glucosylations catalyzed by PlUGT2 in Pueraria lobata. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; CHR, chalcone reductase; IFS, isoflavone synthase; HID, 2-hydroxyisoflavanone dehydratase; PlUGT2, P. lobata UDP-glucosyltransferase 2; IOMT, isoflavone O-methyltranferases. The dotted arrow indicates that the enzymes involved were not yet clear. (1) genistein 4′,7-O-diglucoside; (2) genistin (genistein 7-O-glucoside); (3) sophoricoside (genistein 4′-O-glucoside); (4) liquiritigenin 4′,7-O-diglucoside; (5) neoliquiritin (liquiritigenin 7-O-glucoside); (6) liquiritin (liquiritigenin 4′-O-glucoside); (7) daidzein 4′,7-O-diglucoside; (8) daidzin (daidzein 7-O-glucoside); (9) daidzein 4′-O-glucoside; (10) naringenin 4′,7-O-diglucoside; (11) naringenin 7-O-glucoside; (12) naringenin 4′-O-glucoside; (13) purearin mono-glucoside; (14) ononin (formononetin 7-O-glucoside).
