FIGURE 3.
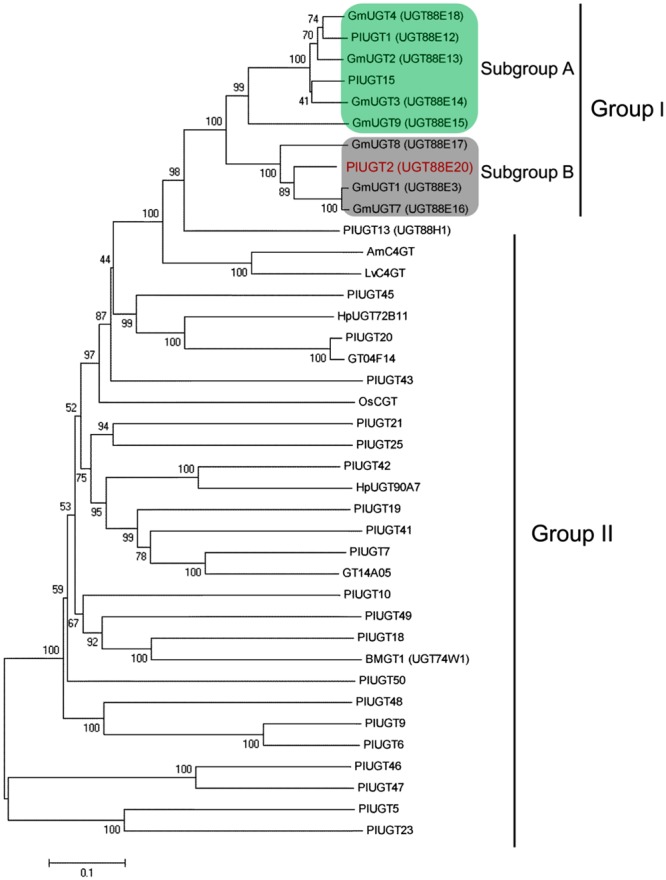
Phylogenetic tree analysis of PlUGT2 with other known UGTs. Twenty two identified P. lobata family 1 UGTs were aligned with other plant UGTs whose functions have been characterized, including seven soybean UGTs and three kudzu UGTs. The tree was constructed from a MEGA 6.0 program using a neighbor-joining method (with 1000 bootstrap replications). Names and accession numbers of UGTs used for the alignment are as follows. AmC4’GT (Antirrhinum majus UDP-glucose: chalcone 4′-O-glucosyltransferase, AB198665); BMGT1 (Bacopa monnieri genistein 4′-O-glucosyltransferase, UGT74W1, ACM09993); GmUGT2 (Glycine max UGT2, AB904891); GmUGT3 (G. max UGT3, AB904892); GmUGT4 (G. max UGT4, AB904893); GmUGT7 (G. max UGT7, AB904894); GmUGT8 (G. max UGT8, AB904895); GmUGT9 (G. max UGT9, AB904896); HpUGT90A7 (Pilosella officinarum flavonoid glucosyltransferases, ACB56926); HpUGT72B11 (P. officinarum coniferyl-alcohol glucosyltransferase, ACB56923); LvC4′GT (Linaria vulgaris UDP-glucose: chalcone 4′-O-glucosyltransferase, BAE48240); OsCGT (Oryza sativa flavoniod C-glucosyltransferase, CAQ77160). PlUGT1 (P. lobata isoflavone 7-O-glucosyltransferase 1, KC473565); PlUGT2 (P. lobata isoflavone 4′,7-O-glucosyltransferase, KU311040); PlUGT13 (P. lobata isoflavone 7-O-glucosyltransferase 13, KC473566); PlUGT15 (P. lobata glucosyltransferase 15, KU311041); PlUGT18 (P. lobata glucosyltransferase 18, KC473567); GT04F14 (P. lobata isoflavone 7-O-glucosyltransferase, HQ219042), GT14A05 (P. lobata flavone glucosyltransferase, HQ219047). The sequences of the other 20 P. lobata glucosyltransferases were submitted to the GenBank database with the accession nos. KU317800–KU317819.
