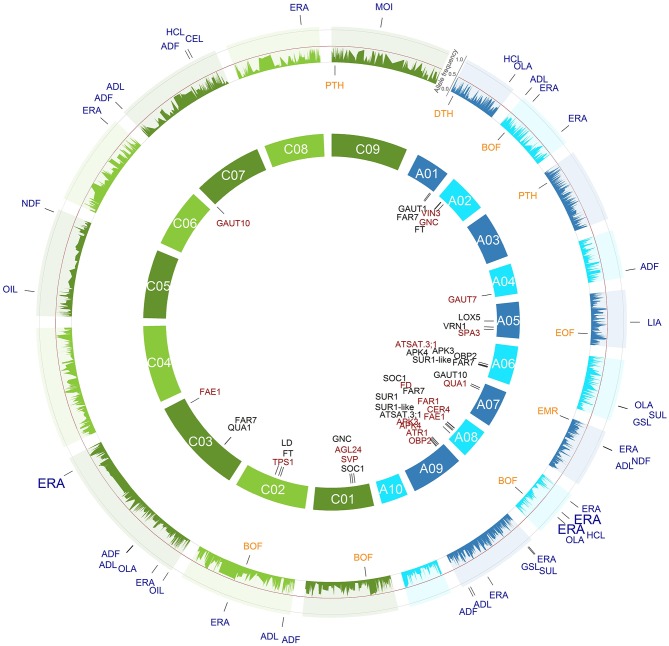
Figure 3.
All single nucleotide polymorphism (SNP)-trait associations with a P < 1.28e-05 (Bonferroni correction of α = 0.05) identified across the 184 inbreds of the spring trial and their respective positions are marked on the B. napus genome. The 3466 SNPs with their minor allele frequencies in the spring trial are given in the outer circle. The SNPs associated with the agronomic SNP-trait associations are plotted in orange below the allele frequency circle and the seed quality SNP-trait associations in blue outside the allele frequency circle. The size of the letters is related to the proportion of the variance explained by the associations. In the inner circle of the 19 chromosomes, the candidate genes were plotted to their mapping position on the B. napus reference genome. The A genome is colored blue and the C genome green.