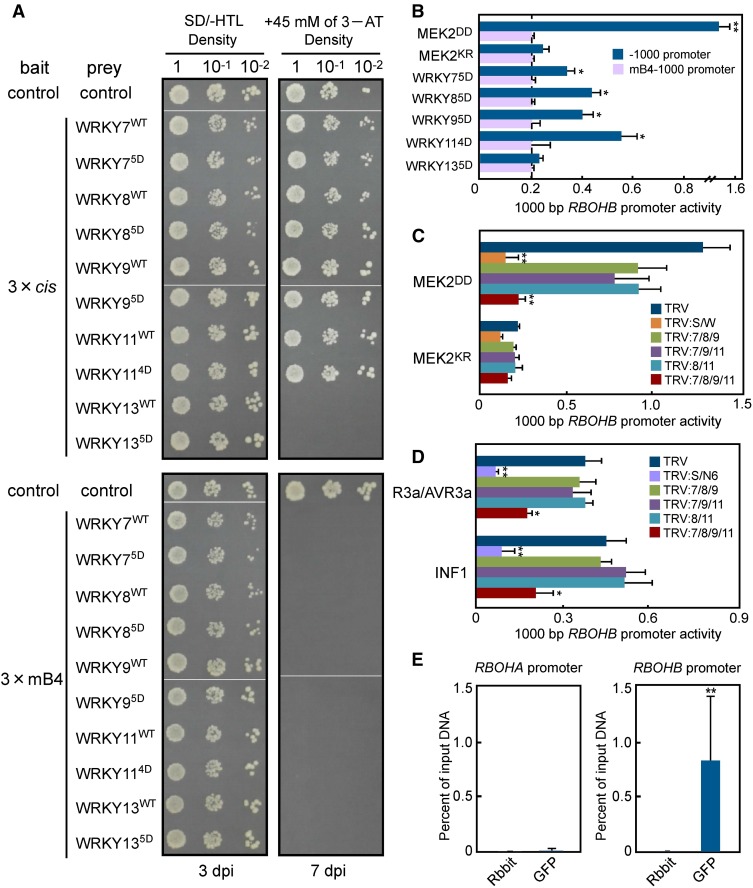
Figure 7.
Involvement of WRKY Transcription Factors in MEK2DD-, INF1-, or R3a/AVR3a-Dependent Activation of the RBOHB Promoter.
(A) Yeast one-hybrid analysis using a three-tandem promoter as bait and WRKY7, 8, 9, 11, or 13 as prey. The representative growth status of yeast cells is shown on SD/-HTL agar media with or without 3-amino-1,2,4-triazole from triplicate independent trials. Numbers on the top of each photograph indicate relative densities of the cells. dpi, days postinoculation.
(B) Analysis of RBOHB −1000 and mB4-1000 promoter in response to WRKY transcription factors. N. benthamiana leaves were inoculated with Agrobacterium carrying promoter assay constructs. WRKY genes were expressed as the effector. Promoter activities were analyzed as described in Figure 2A.
(C) Involvement of multiple WRKY genes in MEK2DD-induced RBOHB promoter activity. Silenced leaves were inoculated with Agrobacterium carrying promoter assay constructs.
(D) Effects of multiple WRKY gene silencing on RBOHB promoter activity induced by INF1 and R3a/AVR3a.
(E) ChIP-qPCR analysis in GFP-WRKY85D-expressed plants. Leaves were inoculated with Agrobacterium carrying pER8:GFP-WRKY85D and then were injected with 20 μM estradiol 24 h later. Input chromatin was isolated from the leaves 12 h after estradiol treatment. GFP-tagged WRKY8-chromatin complex was immunoprecipitated with an anti-GFP antibody. A control reaction was processed at the same time using rabbit IgG. ChIP- and input-DNA samples were quantified by qPCR using primers specific to the promoters of RBOHA and RBOHB genes. ChIP results are shown as percentages of input DNA.
Asterisks indicate statistically significant differences compared with mB4-1000 promoter (B), TRV ([C] and [D]), and rabbit (E) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from three experiments.