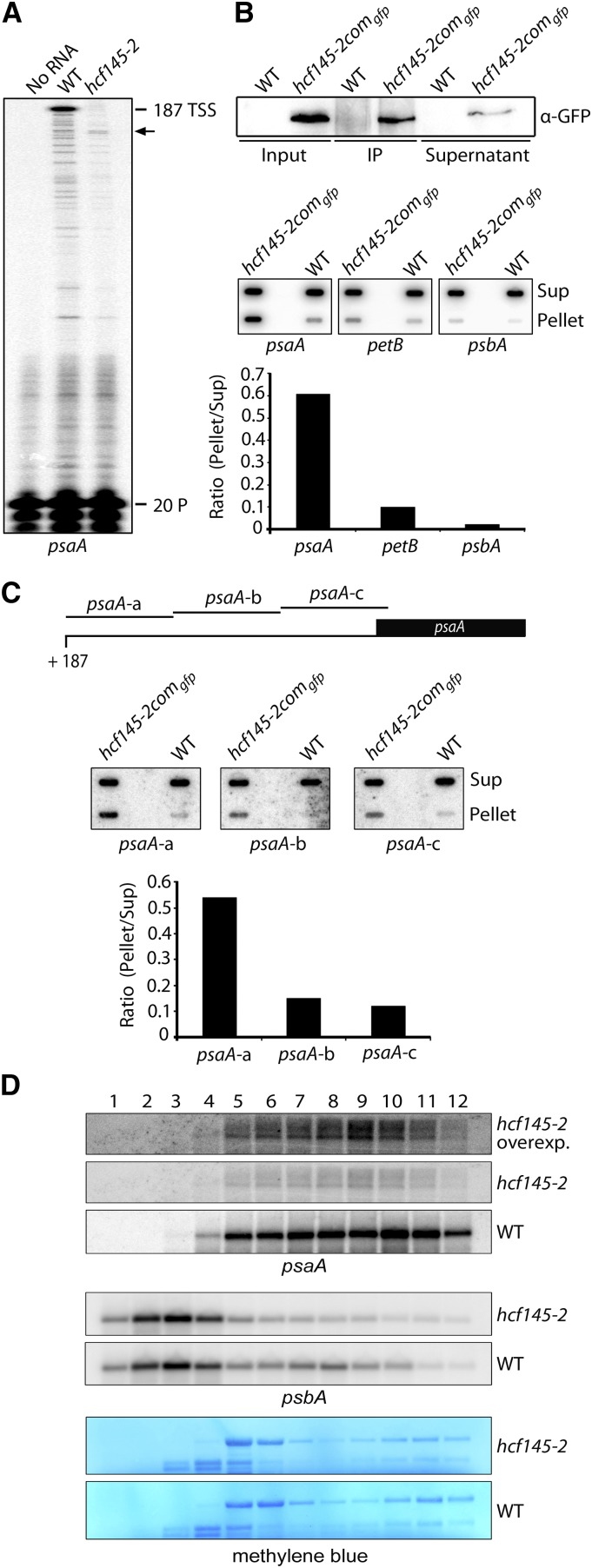
Figure 6.
Determination of the psaA 5′ Termini, RNA Immunoprecipitation, and psaA Polysome Loading of hcf145-2.
(A) Primer extension analysis with radiolabeled primer (20 P) indicates that the 5′ UTR of psaA is truncated in the Arabidopsis hcf145-2 mutant as indicated by arrows. TSS, transcription start site.
(B) Detection of RNA coimmunoprecipitating with HCF145. Upper panel: Immunoprecipitation of the HCF145-GFP fusions from chloroplast extracts of hcf145-2comgfp using GFP antibodies. HCF145-GFP expression was driven by the 35S promoter. IP, immunoprecipitate. Lower panel: Coprecipitated RNAs of the supernatant (Sup) and the pellet were applied to slot blots. Filters were hybridized with the psaA 5′ UTR, petB ORF-, and psbA 5′-region-specific probes. The signals were quantified using ImageQuant software and the ratio of bound versus unbound RNA is indicated in the bar graph.
(C) Fine-mapping of the psaA 5′ UTR RNAs that coimmunoprecipitate with HCF145. Slot-blot hybridization (as in [B]) with three probes covering the entire psaA 5′ UTR. The ratio of bound versus unbound RNA is indicated in the bar graph.
(D) RNA gel blot analysis of polysome fractions 1 to 12 taken from the sucrose gradient of the wild-type and hcf145-2 plants. The probes used are indicated. rRNA was stained with methylene blue. Due to the low expression of psaA in the hcf145-2 mutant, the filter was overexposed (overexp.) for better comparison.