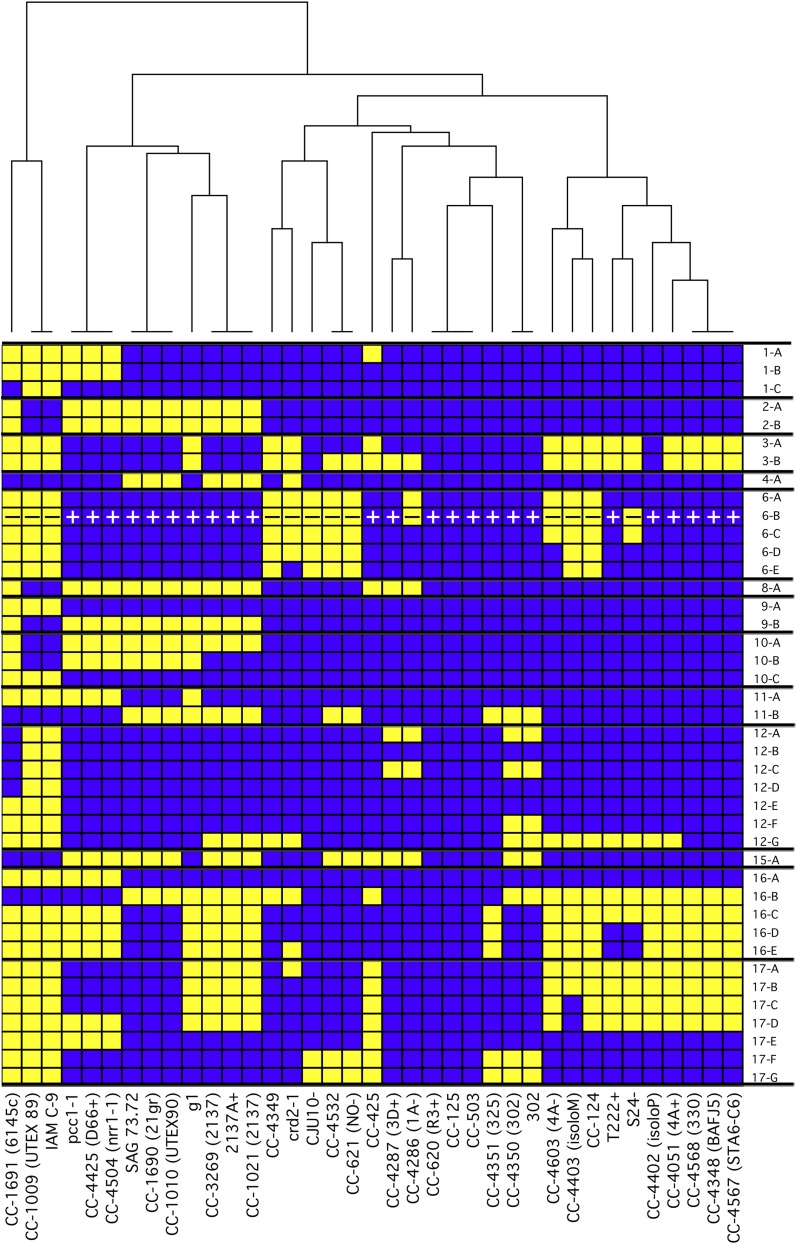
Figure 5.
Distribution of Two Haplotypes in Laboratory Strains.
Within the population of standard laboratory strains, 25.2% of the genome was found to have either of two haplotypes with 2.0% sequence divergence between them. The haplotype of the reference strain, CC-503, was arbitrarily designated as haplotype 1 and the alternate regions as haplotype 2. We algorithmically determined the boundaries of the regions with two haplotypes and combined them into the fewest number of contiguous regions in which all strains are entirely one or the other haplotype. These are plotted for the indicated strains with blue representing haplotype 1 and yellow representing haplotype 2. The mating locus is indicated by + or –. A dendrogram was constructed to arrange the strains based on the similarity of their haplotypes. The coordinates of each block in version 5 of the Chlamydomonas reference genome are presented in Table 2.