Figure 2.
Positional Cloning of the ALIX Gene and the Characteristics of Its Protein.
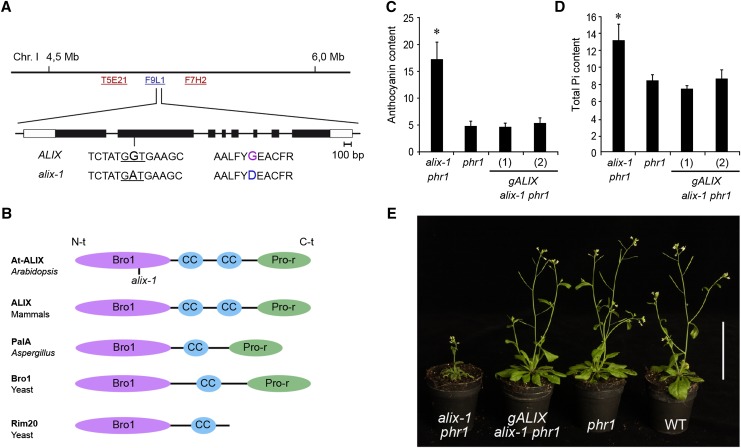
(A) Position of the ALIX locus on chromosome 1 of Arabidopsis (BAC F9L1). The sequence surrounding the alix-1 mutation (G-to-A transition), as well as the resulting amino acid change (Gly260-to-Asp), is shown. The exon structure of ALIX is represented with boxes (light, untranslated; dark, coding region).
(B) Diagrams depicting the domain organization of At-ALIX and homolog proteins in mammals (ALIX), A. nidulans (PalA), and yeast (Bro1 and Rim20). ALIX-related proteins comprise a Bro1 domain (Bro1) in the N-terminal region (N-t), followed by one or two coiled-coil (CC) domains and a proline-rich (Pro-r) motif in the C-terminal region (C-t). The position of the alix-1 mutation in the Bro1 domain of At-ALIX protein is indicated.
(C) and (D) Complementation of alix-1 mutant defects using a construct containing the ALIX genomic region. Plants corresponding to phr1, alix-1 phr1, and alix-1 phr1 transformed with a construct containing the ALIX genomic region (gALIX alix-1 phr1) were grown for 12 d in Pi-deficient medium for anthocyanin measurements (Abs 530 nm/g fresh weight; n = 8) and for 10 d in complete medium for Pi content analysis (μM/g fresh weight; n = 6). Error bars indicate standard deviations. *P < 0.05 (Student’s t test) with respect to the phr1 mutant in the same experimental conditions.
(E) Plants corresponding to the same genotypes as in (C), together with wild-type (WT) plants were grown in soil for 45 d under long-day conditions. Bar = 5 cm.