Figure 3.
ALIX Forms Dimers and Is Essential for Plant Life.
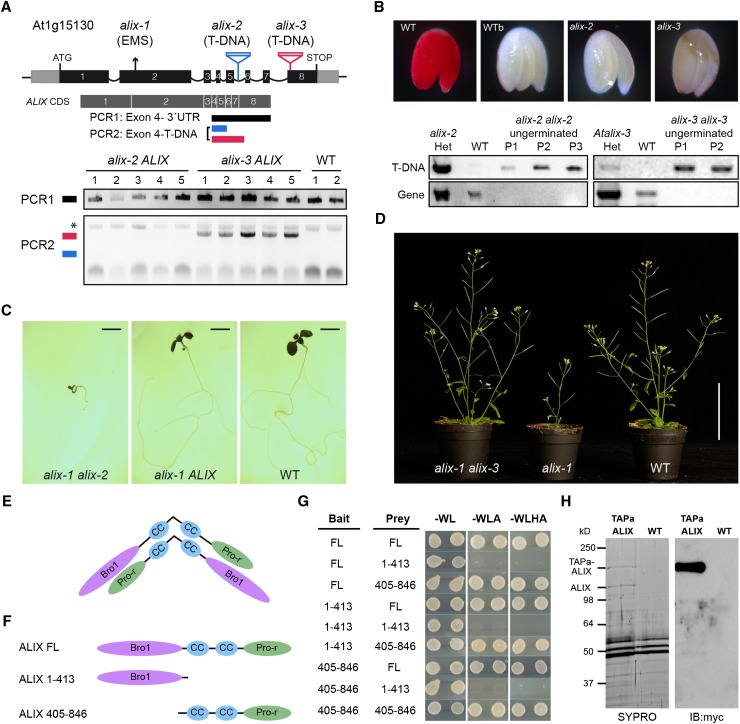
(A) A diagram of At-ALIX genomic region showing the position of the EMS-induced G-to-A mutation in alix-1 plants and that of T-DNA insertions in alix-2 and alix-3 mutants. In lower panels, PCR analysis was used to detect potential truncated ALIX transcripts in the cDNA of alix-2 and alix-3 heterozygous mutants (five lines per mutant). An asterisk indicates a nonspecific band amplified in all samples.
(B) Genotyping and tetrazolium staining shows that embryos from ungerminated seeds of the alix-2 and alix-3 progenies correspond to homozygous mutants that are not viable. Embryos from imbibed (WT) and boiled wild-type (WTb) seeds were used as a control. PCR analysis of wild-type and alix mutant embryos was performed as described in Methods to detect T-DNA insertions in the ALIX gene. P represents pools of 15 embryos.
(C) Photographs of 10-d-old nonviable transheterozygous alix-1 alix-2 and viable heterozygous alix-1 ALIX and wild-type seedlings. Bars = 0.2 cm.
(D) Photographs of trans-heterozygous alix-1 alix-3, homozygous alix-1, and wild-type seedlings grown in soil. Bar = 5 cm.
(E) Model for hypothetical ALIX dimerization following an antiparallel disposition.
(F) and (G) Yeast two-hybrid assays using full-length (FL) and truncated versions (comprising the Bro1 domain, amino acids 1 to 413; or the coiled coils plus the Pro-rich region, amino acids 405 to 846) of ALIX. Transformed yeast cells were grown in SD-WL medium as a transformation control and in SD-WLA and SD-WLHA media for interaction assays.
(H) Copurification of TAPa-ALIX and endogenous ALIX proteins. TAPa-purified proteins were separated in a 10% SDS-PAGE gel and subjected to immunoblot analysis using anti-myc. SYPRO staining was used to visualize differentially purified protein bands prior to their mass spectrometry analysis. Wild-type protein extract was used as a TAPa negative control.