Figure 5.
ERF3 Directly Binds to RR2 in Vitro and in Vivo.
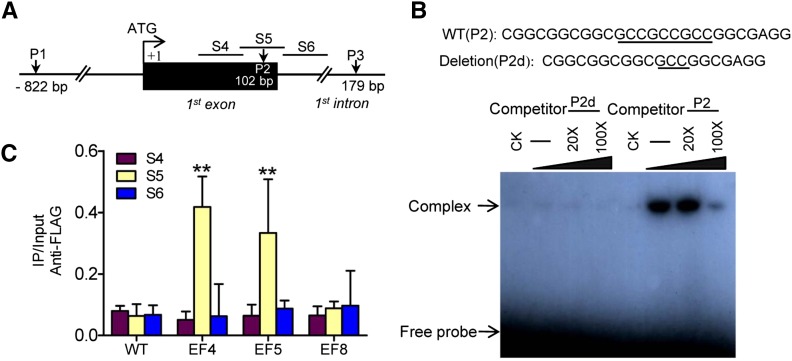
(A) Schematic representation of three putative loci of ERF binding sites in the RR2 genomic sequence. P1 (−822 bp) including GCCGCC motif, P2 (within the first exon) including GCCGCCGCC motif, and P3 (within the first intron) including GCCGCC motif.
(B) Gel-shift assay of ERF3 protein binding to the first exon sequence (P2) of RR2 containing the ERF binding site (underlined) or a deletion version (P2d). E. coli-produced ERF3 protein was incubated with 32P-labeled P2 or P2d in the absence or presence of 20 or 100 M excess of the corresponding cold probes and analyzed by electrophoresis. The shifted band is indicated by the arrow. Three biological replicates were conducted.
(C) ChIP analysis of transgenic plants expressing ERF3-FLAG fusion protein. Nuclei from three ERF3-FLAG transgenic lines (EF4, EF5 and EF8, EF8 as a negative control) and the wild type (WT) were immunoprecipitated by anti-FLAG or without antibody. The precipitated chromatin fragments were analyzed by qPCR using three primer sets amplifying three RR2 regions (S4, S5, and S6), as indicated in (A). The relative nucleotide positions of the putative ERF binding site are indicated (with the initiation ATG codon being assessed as +1). One-tenth of the input chromatin was analyzed as control. Error bars represent means ± sd from three independent experiments.