Fig. 3.
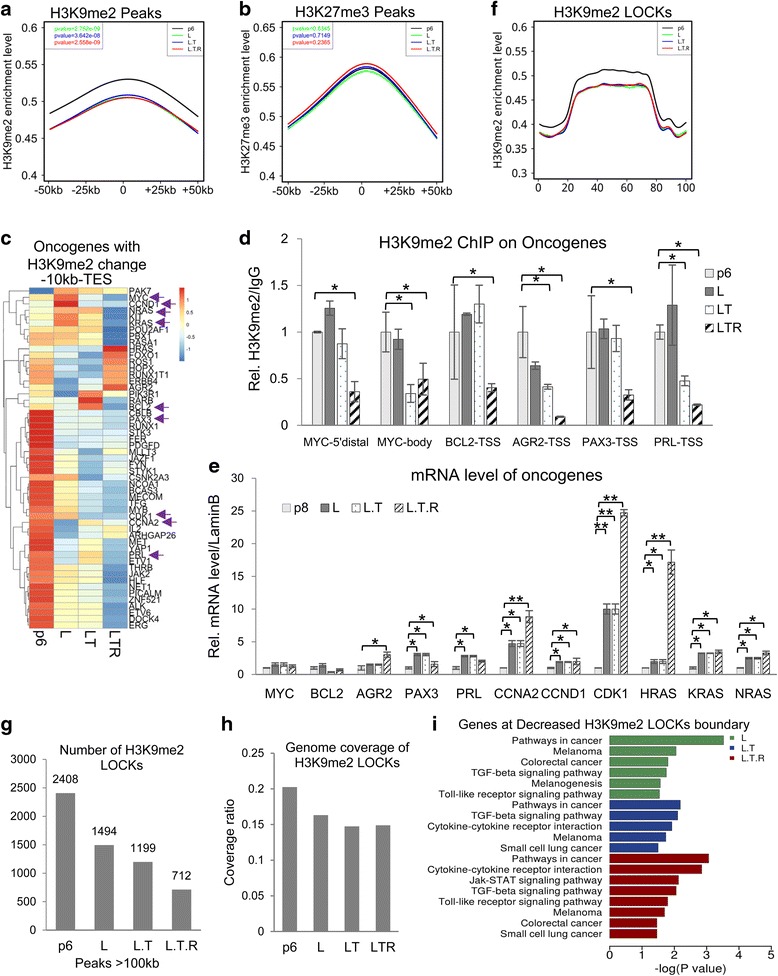
Decreased H3K9me2 alters the transcription program of cancer-related genes at the boundaries of LOCKs. a, b Average enrichment levels of H3K9me2 (a) and H3K27me3 (b) around detected peaks. c The heatmap shows the dynamic H3K9me2 levels on the oncogenes with H3K9me2 reduction. The reads of all H3K9me2 peaks from −10 kb of TSS to TES on each gene were used. The genes verified later by ChIP-PCR and RT-PCR are labelled by arrows. d H3K9me2 levels on several oncogenes were confirmed by ChIP-PCR. e The mRNA expression level of oncogenes with decreased H3K9me2 in transformed cell lines. f The average levels of H3K9me2 LOCKs in the four cell lines. Peaks with length >100 kb were defined as LOCKs. All LOCKs in the four cell lines were extended down- and upstream for 50 kb, and their average H3K9me2 enrichment levels were calculated. g, h The number and average coverage of H3K9me2 LOCK number in four cell lines. i KEGG pathway analysis of genes located at the boundaries of LOCKs with decreased H3K9me2
