Figure 5.
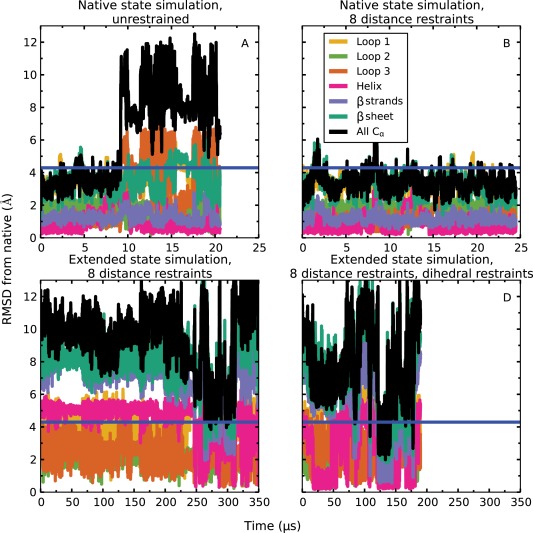
Simulations of domain 1 of CASP target Tc684 (PDB ID: 4GL6). Unrestrained simulations starting from the native state (A) drifted away from the starting conformation, but the native state was stable if the eight native contacts supplied by CASP were enforced using distance restraints (B). A simulation starting from an extended conformation with the distance restraints imposed did not converge to native‐like conformations within 360 µs (C). When dihedral restraints based on predicted secondary structure were also applied (and subsequently released, after formation of the helix, at about 46 µs), a simulation starting from the same extended conformation as in (C) adopted the ensemble realized in (B) between about 120 µs and 140 µs. The blue horizontal line in all plots corresponds to an RMSD of 4.3 Å, which is the lowest RMSD among CASP submissions. Here, “Loop 1” (residues 51–60) is the loop between strand 1 and strand 2, “Loop 2” (residues 70–75) is the loop between strand 2 and strand 3, and “Loop 3” (residues 83–91) is the loop between strand 3 and strand 4. The “β sheet” (residues 44–83) refers to the three major β strands and the two loops between them, whereas “β strands” (all residues in the “β sheet” other than the residues in “Loop 1” and “Loop 2”) refers to just the three major β strands.
