Figure 1.
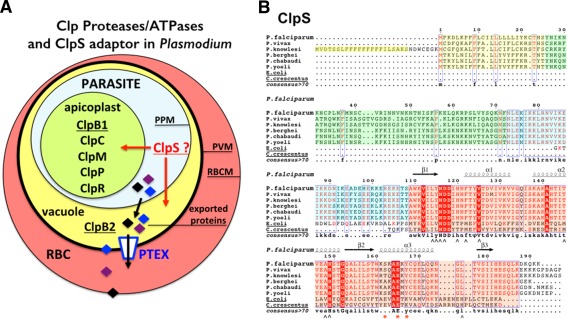
(A) Cellular localization of Clp chaperones and proteases in the parasite Plasmodium falciparum. A red blood cell (RBC) infected by a malaria parasite is shown. Within its host cell, the parasite is self‐contained in the parasitophorous vacuole. PPM, parasitic plasma membrane; PVM, parasitophorous vacuole membrane; RBCM, red blood cell membrane. The apicoplast contains most of the Plasmodium Clp chaperones and proteases (ClpB1, ClpC, ClpM, ClpP, and ClpR) while the ClpB2/Hsp101 protein, a core subunit of the Plasmodium translocon of exported proteins, localizes to the vacuole. No ClpA has been identified in Plasmodium. (B) Sequence alignments of ClpS‐like proteins from different Plasmodium and bacterial ClpS from E. coli and Ccre. In the proteins from the different Plasmodium species, the signal sequences are boxed in yellow. The conserved structural core in all ClpS is boxed in pink. The sequences boxed in green or blue respectively correspond to poorly or highly conserved sequences in Plasmodia; they are part of the predicted apicoplast targeting transit peptide. Secondary structure elements of P. falciparum ClpS are indicated. ∧ symbols indicate residues involved in N‐degron binding in bacterial ClpS structures; red asterisks indicate residues involved in specific binding of ClpS to the N‐terminal domain of ClpA in bacteria.
