Figure 4.
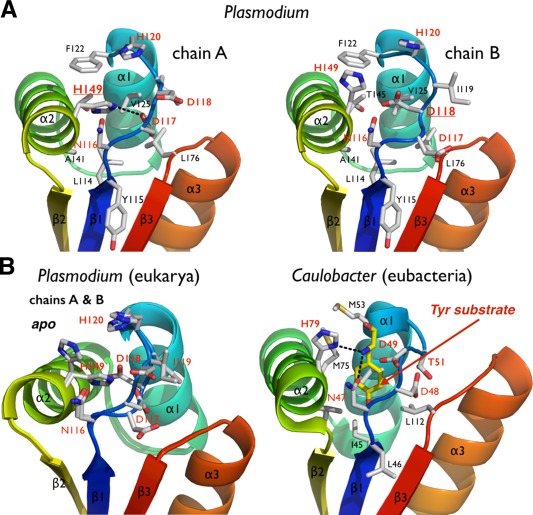
The putative N‐end residue binding site of Plasmodium ClpS. (A) The two copies of ClpS in the asymmetric unit display slightly different NDD loop conformations. (B) Comparison of the Plasmodium and bacterial N‐end degron binding pockets. Equivalent residues are labeled. The bacterial structure (Caulobacter) is shown bound to a tyrosine N‐end rule degron (PDB 3DNJ).11 For Plasmodium, the two chains observed in the asymmetric unit are superposed to emphasize the rigidity of the protein scaffold and the flexibility of the NDD loops.
