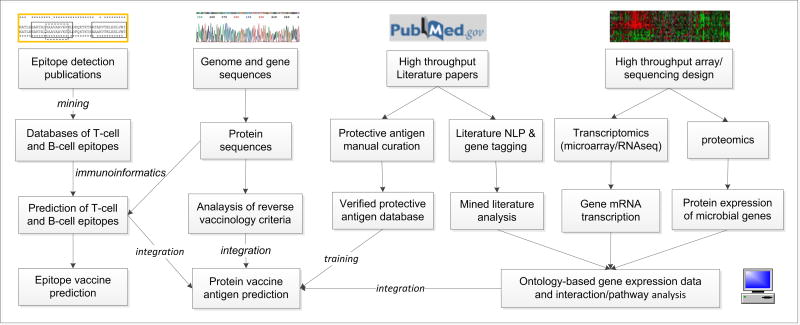
Figure 1. in silico protective vaccine antigen prediction.
Different methods can be used for predicting protective antigens for vaccine development. Specifically, T- and B-cell epitopes can be predicted using protein sequences as input based on training data available from existing immune epitope databases. Bioinformatic analysis of microbial genome sequences allows in silico prediction of protective vaccine antigens using the reverse vaccinology strategy. The database of verified protective antigens (e.g., Protegen) provides gold standard positive antigens for various programs to evaluate. The gene expression data, coming from transcriptomics (DNA microarray or RNA-seq), proteomics, or literature mining can be analyzed and used for further protein vaccine antigen predictions.